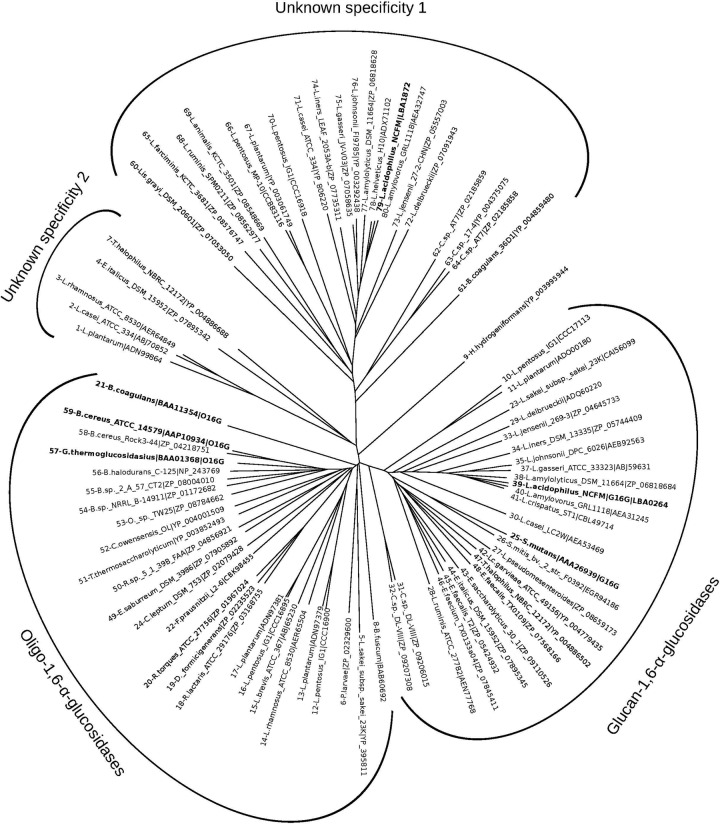
Fig 2.
Phylogenetic tree constructed based on the multiple sequence alignment partly shown in Fig. 1 (full alignment is shown in Fig. S1 in the supplemental material). The tree depicts the clustering of three main groups of GH13 enzymes: (i) a glucan 1,6-α-glucosidase (G16G) cluster from mainly acidophilus complex lactobacilli including LaGH13_31, streptococci, enterococci, and others (see Table S3); (ii) an oligo-1,6-α-glucosidase (O16G) cluster containing characterized enzymes from bacilli, together with a variety of uncharacterized sequences from other Gram-positive bacteria including a branch of non-acidophilus complex lactobacilli; (iii) a cluster of uncharacterized sequences homologous to LBA1872 that occurs in the maltooligosaccharide operon in L. acidophilus NCFM and forms a distinct group (Unknown specificity 1), supportive of the lack of α-1,6 sequence motifs and suggestive of a different function. A small group of uncharacterized sequences form a fourth intermediate group (Unknown specificity 2), whereas a single sequence resembling possibly an ancestral O16G segregates alone, likely due to its taxonomic distance to other O16G sequences in the tree. The following Lactobacillus species are represented: L. amylolyticus, L. animalis, L. brevis, L. delbrueckii, L. farciminis, L. jensenii, L. helveticus, L. iners, L. pentosus, L. plantarum, L. rhamnosus, L. ruminis, and L. sakei subsp. sakei. Other species are as follows: B. halodurans, Bacillus halodurans; B. sp. 2_A_57_CT2, Bacillus sp. strain 2_A_57_CT2; B. sp. NRRL B-14911, Bacillus sp. strain NRRL B-14911; B. fuscum, Brevibacterium fuscum; L. garvieae, Lactococcus garvieae; C. sp. AT7, Carnobacterium sp. strain AT7; C. sp. 17-4, Carnobacterium sp. strain 17-4; C. sp. DL-VIII, Clostridium sp. strain DL-VIII; C. leptum, Clostridium leptum; C. owensensis, Caldicellulosiruptor owensensis; D. formicigenerans, Dorea formicigenerans; E. saccharolyticus, Enterococcus saccharolyticus; E. faecium, Enterococcus faecium; E. italicus, Enterococcus italicus; E. saburreum, Eubacterium saburreum; F. prausnitzii, Faecalibacterium prausnitzii; H. hydrogeniformans, Halanaerobium hydrogeniformans; Lis. grayi, Listeria grayi; L. pseudomesenteroides, Leuconostoc pseudomesenteroides; O. sp. TW25, Ornithinibacillus sp. strain TW25; P. larvae, Paenibacillus larvae; R. sp. 5_1_39B_FAA, Ruminococcus sp. strain 5_1_39B_FAA; R. lactaris, Ruminococcus lactaris; R. torques, Ruminococcus torques; S. mitis, Streptococcus mitis; T. halophilus, Tetragenococcus halophilus; T. thermosaccharolyticum, Thermoanaerobacterium thermosaccharolyticum. Other species are as identified in the text.