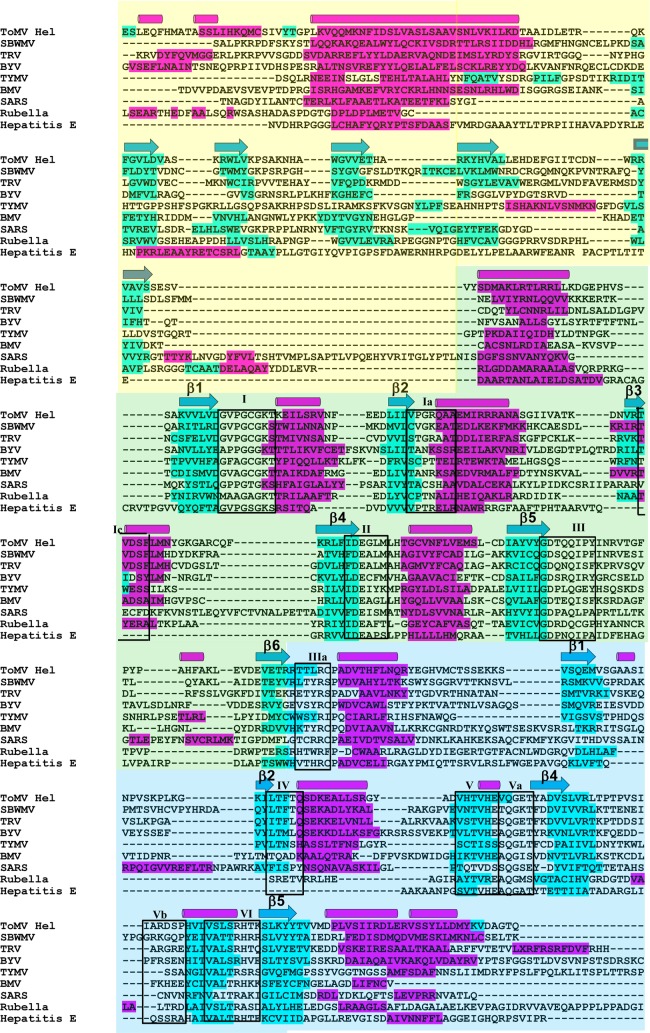
Fig 7.
Sequence alignment of the helicase N-terminal and core domains from the replication proteins of positive-strand RNA viruses. Secondary-structure predictions were made by Jpred (6) for the viral SF1 helicases, and then the nine sequences were manually aligned according to their secondary structures and sequence similarities. The sequences that form β-strands and α-helices in the ToMV-Hel crystal structure are shown above the sequences as blue arrows and magenta cylinders, respectively. The sequences predicted by Jpred to form β-strands and α-helices are highlighted in blue and magenta, respectively. The 11 helicase motifs that are conserved in all of the sequences are boxed. SBWMV, Soilborne wheat mosaic virus (Furovirus, Virgaviridae); TRV, Tobacco rattle virus (Tobravirus, Virgaviridae); BYV, Beet yellows virus (Closterovirus, Closteroviridae); TYMV, Turnip yellow mosaic virus (Tymovirus, Tymoviridae); BMV, Brome mosaic virus (Bromovirus, Bromoviridae); SARS, SARS coronavirus (Coronavirus, Coronaviridae); Rubella, Rubella virus (Rubivirus, Togaviridae); Hepatitis E, Hepatitis E virus (Hepevirus, Hepeviridae).