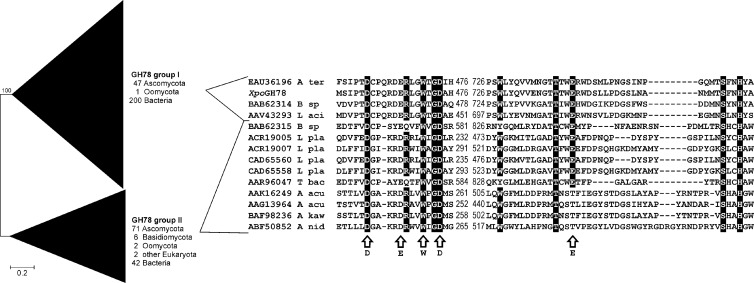
Fig 4.
Phylogenetic analysis of eukaryotic and prokaryotic GH78 α-l-rhamnosidases defining two major groups (I and II). The analysis was performed using the neighbor-joining method with Poisson-corrected protein distances and 500 bootstrap replicates. A total of 371 protein sequences with a total of 3,939 positions were included. The protein alignment comprises two conserved stretches involved in substrate recognition of fungal and bacterial representatives of both main groups as well as the novel characterized X. polymorpha enzyme (XpoGH78). Arrows indicate structural and functional important positions deduced from the three-dimensional structure of Bacillus sp. strain G1 (PDB entry 2OKX; GenBank accession number BAB62315 [11]). A ter, A. terreus; B sp, Bacillus sp.; L aci, Lactobacillus acidophilus; L pla, Lactobacillus plantarum; T bac, Thermomicrobia bacterium; A acu, Aspergillus aculeatus; A kaw, A. kawachii; A nid, A. nidulans.