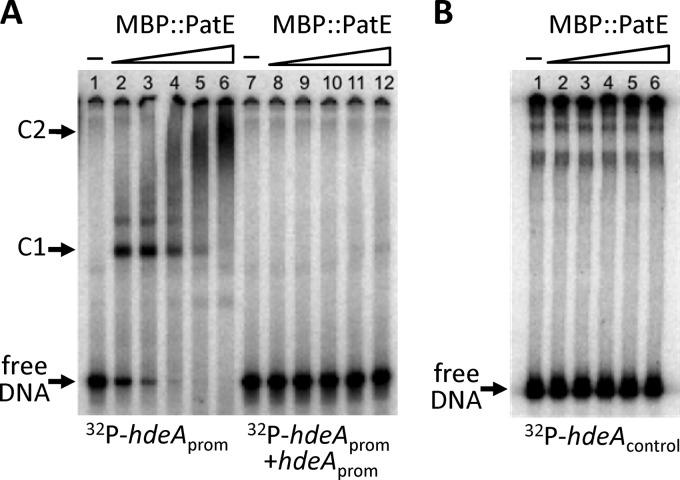
Fig 3.
EMSA analysis of the binding of the fusion protein MBP::PatE to the hdeA promoter region. 32P-labeled PCR fragments were incubated for 30 min at 25°C with increasing amounts of MBP::PatE, after which the samples were analyzed on native polyacrylamide gels. Unbound DNA (free DNA) and DNA-protein complexes (C1 and C2) are indicated. (A) The hdeA promoter fragment (positions −252 to +50 relative to the transcriptional start site) was mixed with 0, 9, 18, 36, 75, or 150 nM MBP::PatE in the absence (lanes 1 to 6) or presence (lanes 7 to 12) of a 100-fold molar excess of an unlabeled hdeA promoter fragment. Densitometric analysis (using the MF-ChemiBIS 3.2 system [DNR Bio-Imaging Systems]) showed that the concentration of MBP::PatE required to shift approximately half of the hdeA fragment (the dissociation constant [Kd]) was 9 nM. (B) A control fragment containing the coding region of hdeA (between positions +162 and +363 relative to the transcriptional start site) was incubated with 0, 9, 18, 38, 75, or 150 nM MBP::PatE.