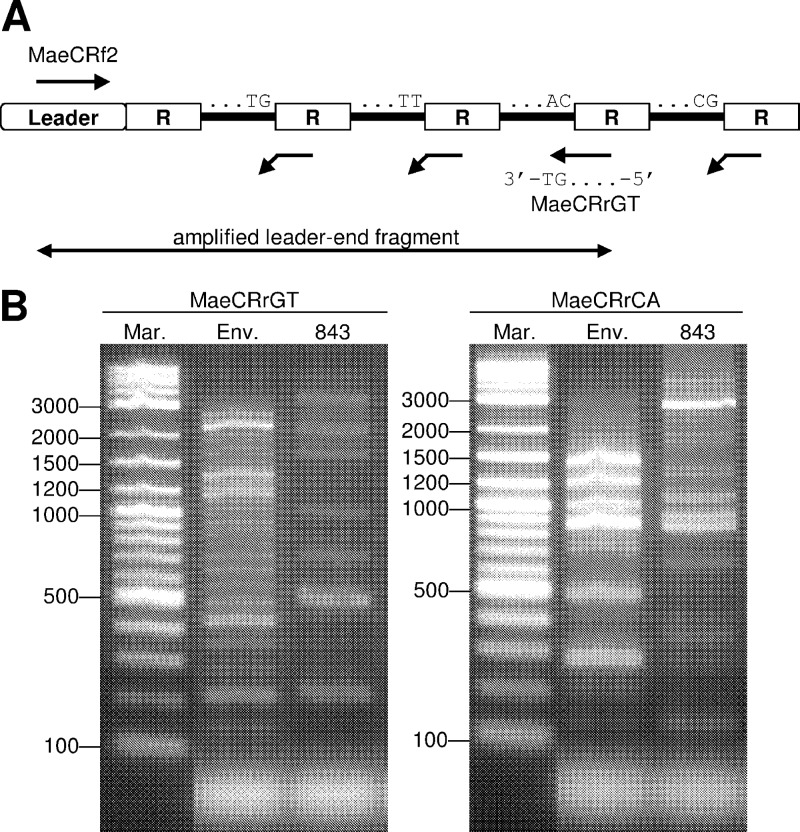
Fig 1.
Amplification of leader-end CRISPR fragments. (A) The reverse primer MaeCRrGT was designed to be complementary to the 5′ end (20 bp) of the repeat (R) but with two additional nucleotides, GT, thereby preferentially annealing to a limited number of specific spacer-repeat units. Another reverse primer, MaeCRrCA, has dinucleotide CA instead of GT. (B) PCR products from primers MaeCRrGT (left) and MaeCRrCA (right). Mar, 2-log DNA ladder; Env, DNA from Hirosawanoike Pond; 843, M. aeruginosa NIES843 genomic DNA.