Abstract
Although the immune response of Caenorhabditis elegans to microbial infections is well established, very little is known about the effects of health-promoting probiotic bacteria on evolutionarily conserved C. elegans host responses. We found that the probiotic Gram-positive bacterium Lactobacillus acidophilus NCFM is not harmful to C. elegans and that L. acidophilus NCFM is unable to colonize the C. elegans intestine. Conditioning with L. acidophilus NCFM significantly decreased the burden of a subsequent Enterococcus faecalis infection in the nematode intestine and prolonged the survival of nematodes exposed to pathogenic strains of E. faecalis and Staphylococcus aureus, including multidrug-resistant (MDR) isolates. Preexposure of nematodes to Bacillus subtilis did not provide any beneficial effects. Importantly, L. acidophilus NCFM activates key immune signaling pathways involved in C. elegans defenses against Gram-positive bacteria, including the p38 mitogen-activated protein kinase pathway (via TIR-1 and PMK-1) and the β-catenin signaling pathway (via BAR-1). Interestingly, conditioning with L. acidophilus NCFM had a minimal effect on Gram-negative infection with Pseudomonas aeruginosa or Salmonella enterica serovar Typhimurium and had no or a negative effect on defense genes associated with Gram-negative pathogens or general stress. In conclusion, we describe a new system for the study of probiotic immune agents and our findings demonstrate that probiotic conditioning with L. acidophilus NCFM modulates specific C. elegans immunity traits.
INTRODUCTION
A number of groups have used the nematode Caenorhabditis elegans to identify and study evolutionarily preserved traits associated with host-pathogen interactions (36). In these C. elegans pathogenesis models, there is significant correlation between the microbial virulence traits required for mammalian infection and the killing of nematodes. In addition, recent studies have described certain evolutionarily conserved defense mechanisms in the nematode, including the insulin/insulin-like growth factor (IGF-1) pathway (28), the p38 mitogen-activated protein kinase (p38 MAPK) pathway (15), the transforming growth factor β (TGF-β) signaling pathway (48), and the β-catenin/HOX transcription factors (12). Therefore, C. elegans has been accepted as an alternative model host for the study of microbial infection and a simple model with which to study evolutionarily conserved aspects of innate immunity (21, 48).
Dietary resources, such as bacteria, play an important role in the control of the C. elegans life span (37), and nematodes exhibit a decreased life span when subjected to a diet of pathogens compared to that when given nonpathogenic laboratory food sources, such as auxotrophic strains of Escherichia coli. Interestingly, it is known that noxious exposure with bacterial pathogens changes C. elegans food preferences (47). In addition to this simple behavior, Anyanful and colleagues (3) showed that preexposure to some pathogenic bacteria can enhance protective responses of C. elegans to a subsequent infection of the pathogen via dopaminergic neurons that activate conserved signaling pathways, a phenomenon they refer to as “conditioning.” However, the challenges with nonpathogenic or health-promoting bacteria and their molecular pathways involved in this phenomenon are poorly understood.
Probiotic bacteria provide functions that are of importance to the health and well-being of the host and contribute in a number of ways toward the functional improvement of foods (16, 45). Interestingly, reports indicate that probiotic bacteria and several lactic acid bacteria (LAB) can enhance the host defense of C. elegans (11, 43). Here we report that the Lactobacillus acidophilus strain NCFM, one of the most widely recognized probiotics that is extensively used in the food and pharmaceutical industries (2), significantly enhances the host defense response of C. elegans to Gram-positive pathogens. The nematode response to conditioning with probiotic bacteria involves a number of previously described host immune responses and is mediated via the TIR-1, PMK-1, and BAR-1 signaling pathways.
MATERIALS AND METHODS
Nematode and pathogenic bacteria.
C. elegans N2 Bristol wild-type, CF512 fer-15(b26)II;fem-1(hc17)IV (fer-15;fem-1 worms), SS104 glp-4(bn2)I,tir-1(qd2) (glp-4 worms), KU25 pmk-1(km25)IV, and EW15 bar-1(ga80)X strains were used in this study. C. elegans strains were routinely maintained on nematode growth medium (NGM) plates seeded with E. coli HB101 using standard procedures (5).
Bacillus subtilis ATCC 6633 (20) and Salmonella enterica serovar Typhimurium SL1344 (1) were cultured on Luria-Bertani (LB) medium at 37°C. Enterococcus faecalis MMH594 (27), E. faecalis V583 (vancomycin-resistant [VRE] clinical isolate) (29), Staphylococcus aureus RN6390 (12), S. aureus MW2 (community-acquired [CA] methicillin-resistant [MRSA] isolate) (4), and Pseudomonas aeruginosa PA14 (15) were grown at 37°C using brain heart infusion (BHI; Difco, Detroit, MI) broth for the E. faecalis strains, tryptic soy broth (TSB; Difco) for the S. aureus strains, and King's broth for P. aeruginosa (38). For liquid killing assays, 50 μl of an overnight culture was spread onto 100 mm BHI agar (for E. faecalis), tryptic soy agar (TSA; for S. aureus assays), or NGM agar (for S. Typhimurium and P. aeruginosa assays) plates. The plates were incubated at 37°C for 18 h and cooled at room temperature (RT) for 1 h (P. aeruginosa for 24 h). For the solid killing assay, 10 μl of E. faecalis cultures was spotted in the middle of BHI agar plates (35 mm) including 80 μg/ml kanamycin, incubated at 37°C for 18 h, and cooled at room temperature for 1 h before beginning the experiment (35). For S. aureus (12), overnight cultures diluted 1:5 in fresh TSB were seeded on TSA plates (35 mm). In addition, S. Typhimurium (39) and P. aeruginosa cultures (22) were seeded on NGM agar plates (35 mm). The plates were incubated at 37°C for 18 h and then cooled at RT for 1 h (P. aeruginosa for 24 h) prior to use.
Preparation of conditioning plates with probiotic bacteria.
To prepare NCFM conditioning plates, L. acidophilus NCFM bacteria (2) were grown in de Man, Rogosa, and Sharpe (MRS) medium (Difco, Detroit, MI) at 37°C for 18 h. Following five washes with M9 medium, 500 μl of each type of cells (ca. 2.0 × 109 CFU/ml) was spread on an NGM plate (100 mm) and dried for 3 h at room temperature. Conditioning plates were stored at 4°C and used within 1 week. E. coli strain HB101 and Bacillus subtilis ATCC 6633 were used as controls, and the same protocols were followed.
To employ heat-killed (HK) cells of the L. acidophilus NCFM strain, overnight cultures were washed twice with sterile M9 medium and resuspended with M9 medium (ca. 2.0 × 109 CFU/ml). Then cells were exposed to 80°C for 60 min, washed with M9 medium, and deposited onto NGM plates. HK cells were plated on MRS to ensure that no viable cells remained.
C. elegans killing assays.
Liquid and solid killing assays were performed using published methods, with slight modifications (5, 35). For the liquid killing assay, L4/young adult worms were placed on conditioning plates with L. acidophilus NCFM at 25°C for 24 h. After exposure to NCFM for 24 h, worms were moved onto lawns of the pathogen and plates were incubated at 25°C for 5 h. Of note is that, as expected, there were no E. coli HB101 bacteria present in the C. elegans gut after they were moved onto an NCFM conditioning plate (data not shown). Then infected worms were washed twice with M9 medium and transferred into wells of a six-well microtiter dish (we used 40 worms per well). Each well contained 2 ml of the liquid assay medium used in previous C. elegans assays (20% BHI and 80% M9) (27). For the solid killing assay, we used 30 worms per plate and worms were transferred to prepared pathogen plates after conditioning on L. acidophilus NCFM plates for 24 h. After worms were placed on the plates with the pathogen, they were incubated at 25°C and examined at 24-h intervals for 15 days for viability using a Nikon SMZ645 dissecting microscope. Worms were counted as alive or dead worms by gentle touching with a platinum wire.
Measuring the bacterial burden in the C. elegans intestinal tract.
The numbers of bacterial cells in the worm intestines were determined according to the modified methods as described previously (8). After conditioning for 24 h, C. elegans animals were moved onto lawns of E. faecalis and incubated at 25°C for 5 h. Worms were washed twice with M9 medium and then transferred into wells of a six-well microtiter dish (we used 40 worms per well). Each worm was removed from the liquid killing plates at 0, 1, 3, and 5 days. After removing surface bacteria, nematodes were washed five times with M9 medium on a BHI plate including gentamicin (25 μg/ml) (8), and worms were placed in new sterile tubes containing M9 medium with 1% Triton X-100 and were mechanically disrupted by using a pestle (Kontes, Vineland, NJ). Persistence was checked by diluting cells by 100 to 107 via 10-fold serial dilution steps in 0.85% NaCl solution, which was plated on BHI agar containing 80 μg/ml kanamycin for E. faecalis or modified MRS (pH 4.5) agar for the L. acidophilus NCFM strain (7). Plates were incubated at 37°C for 24 h. For microscopic observation, at 5 days into the assay, nematodes were quickly placed on a pad of 2% agarose in a drop of 10 mM NaN3 in M9 medium. The worms were examined at ×100 magnification with microscopy.
Induction of the green fluorescent protein (GFP) fusion.
To elucidate the transcriptional host responses [clec-60::GFP in the N2 wild type or clec-60::GFP in the bar-1(ga80) mutant], nematodes were conditioned on plates seeded with L. acidophilus NCFM. After 12 or 24 h, animals were mounted on glass slides with 2% agarose pads, anesthetized with 10 mM NaN3, and quickly visualized using an AxioImager Z1 fluorescence microscope (Zeiss) with A-Plan ×10 magnification. Images were taken using an AxioCam HR camera and analyzed with AxioVision 4.6 (Zeiss) software.
RNA isolation and qRT-PCR.
Total RNA from worms was quickly isolated following the protocol of the TRIzol reagent (Invitrogen) and purified using the RNeasy minikit (Qiagen) including an on-column DNase digestion with RNase-free DNase (Qiagen). After RNA isolation, 50 ng of total RNA was used for a quantitative real-time PCR (qRT-PCR) using the SuperScript III Platinum SYBR green one-step qRT-PCR kit (Invitrogen). qRT-PCR was performed using the CHROMO4 real-time PCR system (MJ Research, Inc., Waltham, MA). Primers were designed using Primer3Input Software (v0.4.0) and are listed in Table S1 in the supplemental material. Relative expression levels were calculated using the 2−ΔΔCT threshold cycle method (23). The control gene snb-1 (12) was used to normalize the genes' expression data.
Statistical analysis.
C. elegans survival was examined by using the Kaplan-Meier method, and differences were determined by using the log-rank test (STATA6; STATA, College Station, TX). Differences in the bacterial numbers from CFU counting were determined by using the Student t test. Each experiment was performed with two different replicates. A P value of 0.05 in all replicate experiments was considered statistically significant.
RESULTS
Probiotic bacteria enhance the resistance of C. elegans to E. faecalis infection.
First, we investigated whether the widely used probiotic bacterium L. acidophilus NCFM (2) influences the life span of C. elegans. As a control, we employed the Gram-positive bacterium B. subtilis strain ATCC 6633 (referred to as B. subtilis 6633), which is nonpathogenic to C. elegans (8). We found no significant difference in the viability of C. elegans exposed to E. coli HB101, B. subtilis 6633, and NCFM in liquid assays with fer-15;fem-1 worms (data not shown) (P = 0.6400 for B. subtilis 6633 and P = 0.2788 for NCFM compared with worms feeding on HB101); hence, we concluded that NCFM is not harmful to C. elegans.
Next we explored if NCFM can augment the C. elegans defense response to E. faecalis. We used both liquid (27) and agar-based solid (35) killing assays. Worms were conditioned by transferring young adult worms to NCFM lawns for 24 h and then transferring them to E. faecalis using liquid and solid killing assays with fer-15;fem-1 worms. Probiotic conditioning with NCFM for 24 h did not affect C. elegans physiological characteristics, including body and brood size, compared to those of worms given the E. coli HB101 control (data not shown). As shown in Fig. 1A, NCFM-conditioned worms clearly exhibited less susceptibility to E. faecalis infection in the liquid killing assay than control worms that were preexposed to the nonpathogenic strain E. coli HB101 (the standard laboratory food source for C. elegans) or the Gram-positive strain B. subtilis ATCC 6633 (P < 0.0001 and P = 0.5762 for NCFM and B. subtilis 6633, respectively, compared to HB101). Consistent with the liquid killing assay, the viability of C. elegans was significantly enhanced when nematodes were conditioned on lawns of NCFM and then exposed to E. faecalis infection in the solid killing conditions (P = 0.0002 and P = 0.0713 for NCFM- and B. subtilis 6633-conditioned nematodes, respectively, compared with worms feeding on HB101) (Fig. 1A). Thus, we concluded that conditioning with the probiotic strain NCFM makes C. elegans worms resistant to E. faecalis and that this beneficial effect was not associated with the preexposure of nematodes to other nonpathogenic bacteria (including the Gram-positive strain B. subtilis 6633).
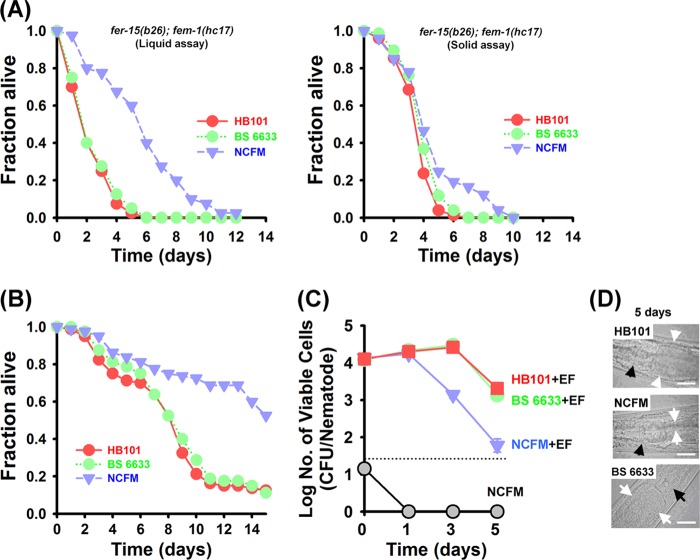
Fig 1.
Immune conditioning with the probiotic bacterium L. acidophilus NCFM prolonged the survival of C. elegans nematodes infected with E. faecalis and decreased the enterococcal burden in the nematode intestine. (A) Liquid (left) (n = 40 per well) and solid (right) (n = 30 per plate) killing assays of C. elegans fer-15;fem-1 worms infected with the E. faecalis MMH594 strain after conditioning with L. acidophilus NCFM or B. subtilis ATCC 6633 for 24 h. Survival statistics: liquid assays, P < 0.0001 and P = 0.5762 for NCFM and B. subtilis 6633, respectively, compared to worms preexposed to E. coli strain HB101; solid assays, P = 0.0200 and P = 0.0713 for NCFM and B. subtilis 6633, respectively, compared to worms preexposed to E. coli strain HB101. (B) Liquid killing assays of C. elegans (n = 40 per well) glp-4 worms infected with the E. faecalis MMH594 strain after conditioning with L. acidophilus NCFM or B. subtilis ATCC 6633 for 24 h. Survival statistics: P < 0.0001 and P = 0.6629 for NCFM and B. subtilis 6633, respectively, compared to worms preexposed to E. coli strain HB101. These findings indicate that the conditioning effect of L. acidophilus NCFM was not specific to the strain of nematodes. (C) Studies evaluating colonization (numbers of CFU/nematode) of L. acidophilus NCFM (gray circles) in the nematode intestine. Also, we illustrate the bacterial burden (numbers of CFU/nematode) of E. faecalis in fer-15;fem-1 nematodes after preexposure to E. coli HB101 (red squares) or conditioning with L. acidophilus NCFM (blue triangles) or B. subtilis ATCC 6633 (green circles). Conditioning with L. acidophilus NCFM significantly inhibited the concentration of E. faecalis in the nematode intestine at day 5. Each experiment was repeated with two independent biological replicates. Error bars show standard deviations. (D) Similar to our findings from the CFU assays, at day 5, the intestinal lumens of worms that had undergone conditioning with L. acidophilus NCFM were slim and without bacteria, while the intestinal lumens of control nematodes that were preexposed to E. coli HB101 or B. subtilis ATCC 6633 were distended by E. faecalis cells. White arrows indicate the borders of the intestinal lumen, and black arrows indicate the grinder organ. Scale bar, 25 μm.
Importantly, this effect was not specific to the strain of nematodes. For example, we employed the well-studied C. elegans fer-15;fem-1 (28) and glp-4 (18) mutants and found that conditioning of worms with NCFM prolonged nematode survival during E. faecalis infection in liquid assays (P < 0.0001 and P = 0.6629 for NCFM- and B. subtilis 6633-conditioned nematodes, respectively, compared with worms feeding on HB101) (Fig. 1B). Also, we performed studies using N2 wild-type nematodes that provided similar results in solid killing assays (data presented below). Unexpectedly, conditioning with heat-killed NCFM bacteria had no effect on resistance to E. faecalis (liquid or solid assays using fer-15;fem-1 worms) (see Fig. S1 in the supplemental material).
Because E. faecalis colonizes the nematode intestinal tract, forming a persistent lethal infection (8, 27), we evaluated the possibility that live NCFM cells colonize the C. elegans intestine, thereby decreasing the number of pathogenic bacteria. In order to investigate this possibility, we evaluated the presence of Lactobacillus and E. faecalis cells in the nematode intestine using CFU studies and microscopic observation. Collaborating with the killing assays, worms were conditioned with NCFM and then exposed to E. faecalis infection. In order to remove surface bacteria, extensive washes were performed on BHI plates containing gentamicin as described previously (8). However, no probiotic bacterial cells could be detected in the nematode intestine during the course of the experiments. In the NCFM-conditioned nematodes, although there was no difference in the number of E. faecalis cells that could be recovered from worms 24 h postconditioning, we found significantly fewer E. faecalis cells in the worm intestine at days 3 and 5 after conditioning than in nonconditioned or B. subtilis-conditioned worms (Fig. 1C). As noted above, we verified these findings using direct microscopic observation. As expected based on the CFU results, worms that were conditioned with NCFM exhibited thin intestinal lumens following infection with E. faecalis, whereas the intestinal lumens of control nematodes preexposed to E. coli HB101 or B. subtilis ATCC 6633 were distended by E. faecalis cells (Fig. 1D). These CFU and microscopy studies confirm previous reports that probiotic bacteria are highly sensitive to passing through the nematode grinder (11). Taken in their totality, these data indicate that probiotic bacteria are digested by the nematode and do not impact the initial colonization of nematodes by E. faecalis. However, conditioning with NCFM significantly inhibited the concentration of E. faecalis in the nematode intestine at 5 days.
Enhanced resistance of C. elegans by conditioning with L. acidophilus NCFM is limited to Gram-positive pathogens.
In the previous sections, we have shown that conditioning with live probiotic bacteria significantly prolongs the survival of C. elegans nematodes during E. faecalis infection and that even though nematodes start with a similar number of pathogenic bacteria, conditioned nematodes are able to decrease the bacterial burden over the course of the infection. These findings raise the hypothesis that conditioning with probiotic bacteria may upregulate nematode host defenses.
Irazoqui and his collaborators found that nematode immune responses to the Gram-negative pathogen Pseudomonas aeruginosa and the Gram-positive pathogen Staphylococcus aureus are distinct (13). More recently, our group found that the nematode responses are also different during fungal infection in whole-transcriptome analysis (33). In order to evaluate the hypothesis that Gram-positive probiotic strains induce species-specific immune responses in C. elegans, we evaluated the impact of conditioning with NCFM on nematode infection with a variety of other Gram-positive and Gram-negative bacteria. We found that, similarly to E. faecalis infection, conditioning with NCFM significantly prolonged the survival of C. elegans exposed to another Gram-positive pathogen, S. aureus, in liquid killing assays with fer-15;fem-1 worms (P = 0.0102 and P = 0.7030 for NCFM and B. subtilis 6633, respectively, compared to nematodes preexposed to E. coli HB101) (Fig. 2A). Similar results were obtained in solid killing assays with N2 wild-type worms (see Fig. 4D). As shown in Fig. 2B, NCFM also prolonged the survival of fer-15;fem-1 animals infected by other enterococcal and staphylococcal strains, such as vancomycin-resistant Enterococcus (VRE) (P = 0.0022 and P = 0.1916 for NCFM and B. subtilis 6633, respectively, compared to nematodes preexposed to E. coli HB101) and methicillin-resistant S. aureus (MRSA) (P = 0.0005 and P = 0.6534 for NCFM and B. subtilis 6633, respectively, compared to nematodes preexposed to E. coli HB101) strains that are among the most commonly recognized multidrug-resistant (MDR) pathogens. However, as noted above, preexposure to the Gram-positive control B. subtilis had no rescuing effect on the survival of nematodes exposed to pathogenic bacteria (Fig. 2A and B).
Fig 2.
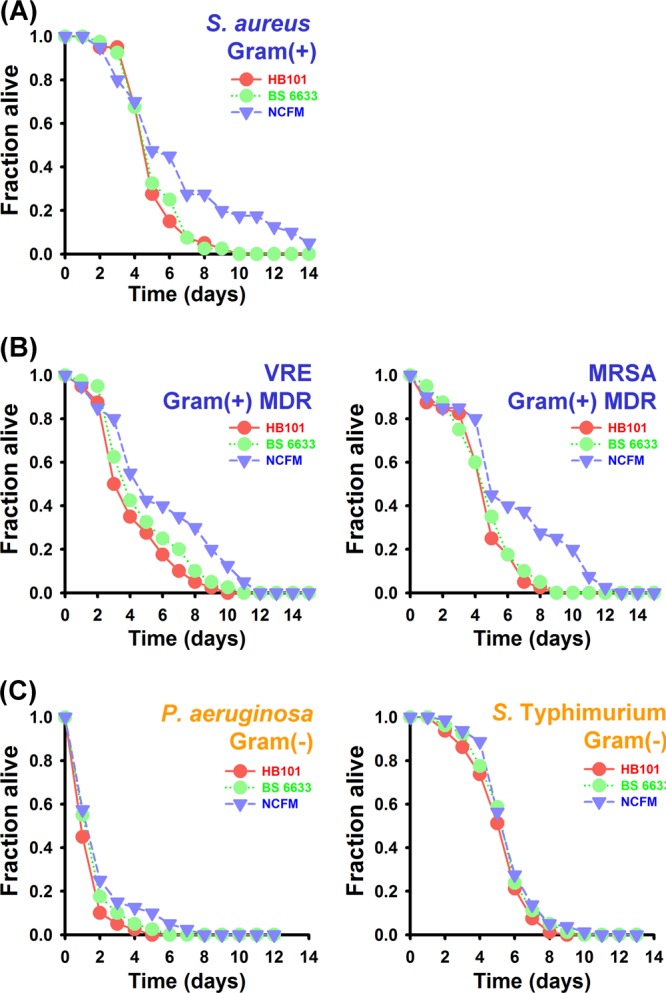
Conditioning with L. acidophilus NCFM selectively influences the resistance of C. elegans worms infected by the Gram-positive bacterium S. aureus but not the Gram-negative pathogens P. aeruginosa and S. Typhimurium. Liquid killing assays with C. elegans fer-15;fem-1 worms (n = 40 per well) infected with S. aureus strain RN6390 (A), E. faecalis strain V583 (VRE strain) or S. aureus strain MW2 (MRSA strain) (B), or P. aeruginosa strain PA14 or S. Typhimurium strain SL1344 (C) following conditioning with L. acidophilus NCFM for 24 h. Survival statistics: S. aureus RN6390, P = 0.0102 and P = 0.7030 for NCFM and B. subtilis 6633, respectively, compared with control worms preexposed to E. coli strain HB101; E. faecalis V583, P = 0.0022 and P = 0.1916 for NCFM and B. subtilis 6633, respectively, compared with control worms preexposed to E. coli strain HB101; S. aureus MW2, P = 0.0005 and P = 0.6534 for NCFM and B. subtilis 6633, respectively, compared with control worms preexposed to E. coli strain HB101; P. aeruginosa PA14, P = 0.0585 and P = 0.2320 for NCFM and B. subtilis 6633, respectively, compared with worms feeding on HB101; and S. Typhimurium SL1344, P = 0.0668 and P = 0.2217 for NCFM and B. subtilis 6633, respectively, compared with worms feeding on HB101.
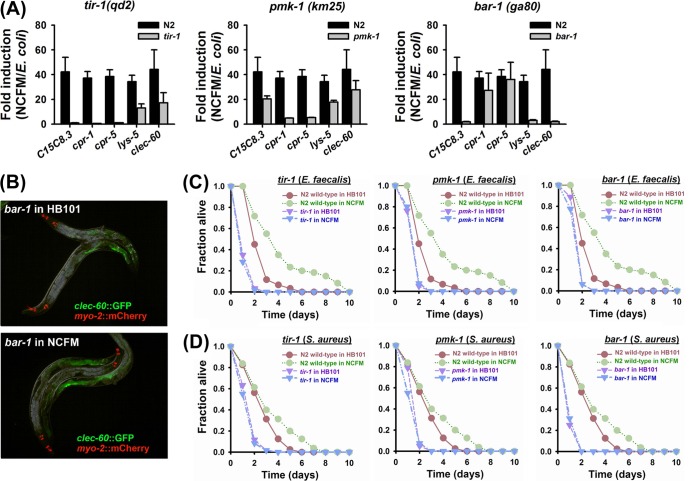
Fig 4.
tir-1, pmk-1, and bar-1 are required for the immune induction by L. acidophilus NCFM. (A) qRT-PCR analysis of five antimicrobial genes (C15C8.3, cpr-1, cpr-5, lys-5, and clec-60) in N2 wild-type and mutant animals after conditioning with L. acidophilus NCFM for 12 h. (B) Limited induction of clec-60::GFP by conditioning with L. acidophilus NCFM in the bar-1 mutant. clec-60::GFP transgenic animals in the wild-type N2 (top) or bar-1 mutant (bottom) background underwent conditioning with L. acidophilus NCFM for 12 h. Images were taken at the same time point with the same exposure time. myo-2::mCherry shows red pharyngeal expression as a marker for the presence of the transgene. (C) Solid killing of C. elegans strain N2 wild-type worms and tir-1, pmk-1, or bar-1 loss-of function mutants (n = 30 per plate) infected by E. faecalis MMH594. Survival statistics: tir-1, P < 0.0001 for tir-1 control worms preexposed to E. coli HB101 and P < 0.0001 for tir-1 worms conditioned with NCFM compared to N2 wild-type worms preexposed to E. coli HB101, P = 0.3843 for tir-1 worms conditioned with NCFM compared to tir-1 worms preexposed to E. coli HB101; pmk-1, P < 0.0001 for pmk-1 worms preexposed to E. coli HB101 and P < 0.0001 for pmk-1 worms conditioned with NCFM compared to N2 wild-type worms preexposed to HB101, P = 0.4326 for pmk-1 worms conditioned with NCFM compared with pmk-1 worms preexposed to HB101; and bar-1, P < 0.0001 for bar-1 after preexposure to E. coli HB101 and P < 0.0001 for bar-1 worms conditioned with NCFM compared to N2 wild-type worms preexposed to E. coli HB101, P = 0.1403 for bar-1 worms conditioned with NCFM compared to bar-1 worms preexposed to E. coli HB101. A 12-h conditioning protocol was used in all studies. (D) Solid killing of C. elegans strain N2 wild-type worms and tir-1, pmk-1, or bar-1 loss-of function mutants (n = 30 per plate) infected by S. aureus RN6390. Survival statistics: tir-1, P < 0.0001 for tir-1 control worms preexposed to E. coli HB101 and P < 0.0001 for tir-1 worms conditioned with NCFM compared to N2 wild-type worms preexposed to E. coli HB101, P = 0.3636 for tir-1 worms conditioned with NCFM compared to tir-1 worms preexposed to E. coli HB101; pmk-1, P < 0.0001 for pmk-1 worms preexposed to E. coli HB101 and P < 0.0001 for pmk-1 worms conditioned with NCFM compared to N2 wild-type worms conditioned with NCFM, P = 0.0225 for pmk-1 worms conditioned with NCFM compared to pmk-1 worms preexposed to E. coli HB101; and bar-1, P < 0.0001 for bar-1 worms preexposed to E. coli HB101 and P < 0.0001 for bar-1 worms conditioned with NCFM compared to N2 wild-type worms preexposed to E. coli HB101, P = 0.4909 for bar-1 worms conditioned with NCFM compared to bar-1 worms preexposed to E. coli HB101. A 12-h conditioning protocol was used in all studies.
On the other hand, conditioning with NCFM had no effect on survival following infection with the Gram-negative pathogen P. aeruginosa in either liquid (Fig. 2C) (P = 0.0585 and P = 0.2320 for NCFM and B. subtilis 6633, respectively, compared with worms preexposed to HB101) or solid (data not shown) (P = 0.5761 and P = 0.8285 for NCFM and B. subtilis 6633, respectively, compared with worms fed HB101) killing assays. In addition, when challenged with another Gram-negative bacterium, S. enterica serovar Typhimurium, similar to challenge with P. aeruginosa, conditioning with NCFM had no statistically significant effect on the survival of C. elegans in either liquid (Fig. 2C) (P = 0.0668 and P = 0.2217 for NCFM and B. subtilis 6633, respectively, compared with worms preexposed to HB101) or solid (data not shown) (P = 0.0826 and P = 0.4811 for NCFM and B. subtilis 6633, respectively, compared with worms preexposed to HB101) killing assays with fer-15;fem-1 worms. These results suggest that the immune induction by the probiotic bacterium strain NCFM may be specific to Gram-positive pathogens.
L. acidophilus NCFM regulates specific gene transcriptions.
We next sought to determine the mechanism associated with C. elegans conditioning with NCFM. Based on our findings that preexposure to the probiotic NCFM has an effect on Gram-positive bacteria but not S. enterica Typhimurium or P. aeruginosa (Gram-negative model pathogens that have been extensively studied in C. elegans), we postulated that conditioning with probiotic bacteria upregulates specific immune factors that are associated with C. elegans infection with E. faecalis or S. aureus. We mined the previous whole-transcriptome results linked to C. elegans infection with E. faecalis and S. aureus (12, 46), looking for upregulated genes with a >4.0-fold increase. We selected 14 target/candidate genes and categorized them in the following three groups: (i) E. faecalis-specific genes, such as asp-1, C15C8.3 (encodes aspartyl proteases), cpr-1 (encodes a cysteine protease), and thn-2 (thaumatin/PR-5), (ii) genes upregulated during both E. faecalis and S. aureus infection, including cpr-4 and cpr-5 (encode cysteine proteases), lys-5 (encodes a lysozyme), and fmo-2 (encodes a flavin-containing monooxygenase), and (iii) S. aureus-specific genes like cpr-2 (encodes a cysteine protease), clec-52, clec-60, and clec-71 (that encode C-type lectins), ilys-3 (encodes a invertebrate lysozyme), and F53A9.8 (encodes a short His-rich protein). We investigated the regulation of these genes in C. elegans nematodes exposed to NCFM using quantitative real-time PCR (qRT-PCR). We found that almost all selected genes were upregulated by >5.0-fold by the strain NCFM. More specifically, our qRT-PCR results indicated that conditioning with NCFM dramatically induced the transcription of C15C8.3 (34.1-fold ± 7.9-fold), cpr-1 (24.6-fold ± 3.1-fold), cpr-5 (38.6-fold ± 7.4-fold), lys-5 (26.5-fold ± 3.9-fold), and clec-60 (46.7-fold ± 7.2-fold) (Fig. 3A). These findings are consistent with our killing assay results that NCFM stimulated the resistance of C. elegans to infection by Gram-positive bacteria (Fig. 1 and 2). We also studied the induction of clec-60 using transgenic animals carrying the clec-60::GFP reporter line (12). Consistent with the qRT-PCR results, we noted strong induction of clec-60::GFP after conditioning with NCFM (Fig. 3B). Consistent with our survival assays (Fig. 1 and 2), preexposure to B. subtilis clearly did not influence the expression of any of the five selected host defense genes (Fig. 3C).
Fig 3.
Conditioning with L. acidophilus NCFM regulates the transcription of immune genes. (A) qRT-PCR analysis evaluating the impact of conditioning on genes associated with nematode immune responses to E. faecalis, S. aureus, and P. aeruginosa as well as stress-related genes (nematodes “conditioned” with normal food, E. coli HB101, were used for comparison). Transcript levels were measured in young adult fer-15;fem-1 worms conditioned with L. acidophilus NCFM for 24 h. (B) Induction of clec-60::GFP exposed to L. acidophilus NCFM for 24 h. myo-2::mCherry shows red pharyngeal expression as a marker for the presence of the transgene. Images were taken at the same time point with the same exposure time. DIC, differential interference contrast. (C) Transcripts of 5 selected genes after conditioning with L. acidophilus NCFM or B. subtilis ATCC 6633 for 24 h. Preexposure to B. subtilis had no impact on the regulation of five selected host defense genes.
Since NCFM did not significantly protect nematodes from infection with P. aeruginosa, we also determined whether NCFM activates the expression of defense-related genes activated by P. aeruginosa infection. Consistent with our results from the killing assays (Fig. 2C), six genes (T24B8.5, C32H11.1, C32H11.12, F35E12.5, C17H12.8, and K08D8.5) predicted to be strongly induced by P. aeruginosa (31, 35, 41) were either unchanged (C32H11.1) or repressed (T24B8.5, C32H11.12, F35E12.5, C17H12.8, and K08D8.5) by NCFM conditioning (Fig. 3A). Notably, the transcription of T24B8.5, which encodes a ShK-like toxin peptide that is regulated by the PMK-1 signaling pathway and the ATF-7 transcription factor when worms are infected by P. aeruginosa (35), was decreased by 15.6-fold ± 3.0-fold. In addition, six general-stress-associated genes (10, 44), sod-3, sod-5, mtl-1, hsp-16.2, old-1, and gst-10, were not affected in the presence of NCFM (Fig. 3A). Taken in their totality, these results indicate that conditioning with the probiotic bacterium NCFM specifically stimulated the transcription of host defense genes associated with nematode responses to Gram-positive pathogens but had no or even a negative effect on genes associated with Gram-negative bacteria or genes involved in the response to general stress.
The nematode host response to conditioning with NCFM is multifactorial and involves the TIR-1, PMK-1, and BAR-1 signaling pathways.
To elucidate the immune pathways stimulated by NCFM, we employed loss-of-function tir-1, pmk-1, or bar-1 C. elegans immune-related mutants and investigated the regulation of the selected genes, including C15C8.3, cpr-1, cpr-5, lys-5, and clec-60, in these mutant strains after conditioning with NCFM. The bar-1 loss-of-function mutation strongly affected the fold induction of clec-60, whereas either the tir-1 or pmk-1 loss-of-function mutation significantly attenuated the upregulation of cpr-1 and cpr-5 (Fig. 4A). Moreover, we observed that clec-60::GFP was not induced by conditioning of the bar-1 mutant with NCFM (Fig. 4B), in contrast to conditioning of the wild type (Fig. 3B). Interestingly, we found that mRNA levels of C15C8.3 and lys-5 were significantly affected by the deletion of tir-1, pmk-1, or bar-1 when they were exposed to NCFM (Fig. 4A); hence, we suggest that the TIR-1, PMK-1, and BAR-1 signaling pathways participate in the regulation of C15C8.3 and lys-5 by NCFM exposure.
Taken in their totality, our results described above support the hypothesis that the TIR-1, PMK-1, and BAR-1 signaling pathways are involved in immune stimulation by conditioning with NCFM. In order to further investigate this hypothesis, we carried out solid killing assays with mutants or N2 wild-type worms to confirm that the TIR-1, PMK-1, and BAR-1 signaling pathways are directly involved in C. elegans by conditioning with NCFM. Surprisingly, worms with deletion of either tir-1, bar-1, or pmk-1 were highly susceptible to E. faecalis and S. aureus. Similar to our results with other nematode strains (Fig. 1 and 2), NCFM significantly induced the resistance of wild-type nematodes (N2 strain) infected by E. faecalis and S. aureus (P < 0.0001 and P = 0.0041, respectively, compared with worms feeding on HB101). However, consistent with qRT-PCR results, worms lacking tir-1, pmk-1, or bar-1 were still susceptible to E. faecalis (Fig. 4C) and S. aureus (Fig. 4D) infections even though they were conditioned with NCFM. Taken together, our results indicate that the TIR-1, PMK-1, and BAR-1 pathways all play an important role in immune conditioning with these probiotic bacteria.
DISCUSSION
Since Metchnikoff's work (26), a wealth of data have shown that probiotic bacteria are involved in host immune responses in humans (9, 24). Probiotic bacteria constantly exist in the human intestinal tract (34), and accumulating evidence indicates that they are involved in the regulation of the host immune system by modulating key signaling pathways (including MAPK, Akt/phosphoinositide 3-kinase [PI3K], and peroxisome proliferator-activated receptor gamma [PPARγ]) and by influencing downstream pathways (40). Because some of these signaling pathways are highly conserved from C. elegans to humans, the question that arises is whether probiotic bacteria have a similar effect in the immunity of C. elegans (14). In this work, we employed a conditioning concept to study the effects of probiotic bacteria in the immune response of C. elegans. Our findings demonstrate that nematode responses can undergo conditioning by probiotic bacteria, making them more resistant to virulent pathogens. We demonstrate that conditioning with the probiotic bacterium L. acidophilus strain NCFM stimulates C. elegans immune responses, making nematodes more resistant to infection by the Gram-positive pathogenic bacteria E. faecalis (Fig. 1) and S. aureus (Fig. 2A), including even antibiotic-resistant strains (Fig. 2B). Importantly, probiotic NCFM bacteria significantly inhibited the burden of E. faecalis in the nematode intestine even though they are completely digested by the nematode (Fig. 1C). In addition, based on C. elegans survival rates and qRT-PCR results, preexposure to the Gram-positive bacterium B. subtilis (Fig. 1, 2, and 3C) did not have the similar beneficial effects, indicating that the conditioning effect we found using the probiotic bacterium NCFM is, at least to some degree, specific.
Our findings on the ability of C. elegans nematodes to undergo conditioning are supported by the work of Anyanful and coworkers (3). These workers demonstrated that preexposure to the avirulent enteropathogenic E. coli (EPEC) ΔtnaA mutant (that is lacking the activity of tryptophanase) can induce responses associated with C. elegans infection with virulent EPEC and that these effects depend on dopaminergic neuronal signaling that activates the insulin/IGF-1 and p38 MAPK pathways (3). Notably, it has been known that cell wall molecules (CWMs) may be key probiotic ligands that interact with host receptors and induce signaling pathways, resulting in immune-stimulating effects (19). Taken along with our findings, it is reasonable to assume that virulent, avirulent, and probiotic microbes share common features, including pathogen- and microbe-associated molecular patterns (PAMPs/MAMPs) such as peptidoglycan, lipoteichoic acid, and polysaccharides (19, 25), and regulate similar nematode immune responses. Equally importantly, the additional possibility is that probiotic bacteria may lead to release or exposure of damage-associated molecular patterns (DAMPs). DAMPs are host-derived molecules that can activate immune responses (42). It is possible that NCFM conditioning triggers the production of DAMPs that mediate responses to Gram-positive bacteria. This hypothesis is also supported by our finding that the immune responses are specific and that NCFM conditioning had no effect on or even resulted in downregulation of genes associated with the nematode responses to the Gram-negative pathogens P. aeruginosa and S. Typhimurium. Of note is that some cross-responses to different DAMPs might exist, as suggested by the experiments by Ikeda et al. in which conditioning with lactic acid bacteria prolonged the survival of nematodes exposed to Salmonella spp. (11). In addition, probiotic effects in human health are multifactorial and they are dependent on the species and strain (19, 45). Ikeda and colleagues (11) employed different probiotic strains, including Bifidobacterium, Lactobacillus helveticus, and Lactobacillus plantarum, in C. elegans studies, whereas we used L. acidophilus NCFM, one of the most widely recognized probiotic bacteria that is extensively used in the food and pharmaceutical industries (2). Thus, probiotic specificity may result in a unique immune-stimulating effect in C. elegans. Indeed, a number of immune-stimulating molecules, including peptidoglycan (19), S-layer protein (18), and exopolysaccharide (EPS) (16, 17), were identified from the probiotic cell surface. Taken together, we conclude that activation of C. elegans host immunity by probiotic bacteria can be mediated by either exogenous (heat-labile PAMPs/MAMPs) or endogenous (DAMPs) features (or both) and that further work is required to elucidate which scenarios/mechanisms are directly involved in probiotic bacterium-mediated host response.
Several immune response signaling pathways have been identified in C. elegans (12, 15, 28, 31, 32, 48). Importantly, we found that C. elegans conditioning with the NCFM probiotic involves the p38 MAPK (via PMK-1) pathways, including the Toll/interleukin-1 receptor (TIR-1) pathway as well as the β-catenin (via BAR-1) signaling pathway (Fig. 4). PMK-1 and BAR-1 are key features of C. elegans innate immunity (14). The previous challengers indicated that either the pmk-1 or bar-1 signaling pathway is strongly involved in infection with the Gram-positive pathogen S. aureus (12). Also, the Toll-like receptor (TLR) signaling pathway is required for the C. elegans immune response to microbial pathogen infection (32). In the C. elegans genome, the following two genes encode TLR-related proteins: (i) tir-1, which encodes a conserved TIR domain protein that is homologous to the mammalian gene known as the sterile-α and Armadillo motif (SARM) (22), and (ii) tol-1, which encodes a potential Toll receptor homologue (32). tir-1 functions upstream of a conserved p38 MAPK cascade and is a positive regulator of PMK-1 (6). Consistent with our results (Fig. 4), inactivation of tir-1 results in enhanced susceptibility of C. elegans to infection with the Gram-positive pathogen E. faecalis (22). Interestingly, this pathway is also part of the mammalian immune response to probiotics (30). Notably, although tol-1 appears to play a role in the nematode response to the Gram-negative bacterium S. Typhimurium, it is not involved in the immune response to the Gram-positive pathogens S. aureus and E. faecalis (13, 39) and C. elegans lacks myeloid differentiation primary response protein 88 (MYD88) and a nuclear factor-κB (NF-κB)-like transcription factor (14). Therefore, we propose that NCFM-mediated conditioning of C. elegans is mediated by multiple pathways, including signaling through PMK-1/TIR-1 and BAR-1.
Taken in their totality, the signaling pathways through PMK-1 (with TIR-1) and BAR-1 are key components of the C. elegans immune conditioning with the probiotic bacterium NCFM and these pathways appear to act in parallel in order to promote immunity. Our working model (see Fig. S2 in the supplemental material) proposes a novel approach to probiotic-mediated immune conditioning in C. elegans. These findings demonstrate that C. elegans immunity can undergo conditioning and that Gram-positive pathogens and probiotic bacteria share key immune response signaling pathways.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from Chonbuk National University (2012) to Y.K. and the NIH (P01 AI083214) to E.M.
We are grateful for the bar-1(ga80) mutants provided by Javier E. Irazoqui. We thank Frederick M. Ausubel for critical comments and discussions and Annie L. Conery and Jonah Larkins-Ford for technical advice.
We declare no conflict of interest.
Footnotes
Published ahead of print 14 May 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Aballay A, Yorgey P, Ausubel FM. 2000. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr. Biol. 10:1539–1542 [DOI] [PubMed] [Google Scholar]
- 2. Altermann E, et al. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. U. S. A. 102:3906–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anyanful A, Easley KA, Benian GM, Kalman D. 2009. Conditioning protects C. elegans from lethal effects of enteropathogenic E. coli by activating genes that regulate lifespan and innate immunity. Cell Host Microbe 5:450–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baba T, et al. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827 [DOI] [PubMed] [Google Scholar]
- 5. Breger J, et al. 2007. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog. 3:e18 doi:10.1371/journal.ppat.0030018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Couillault C, et al. 2004. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 5:488–494 [DOI] [PubMed] [Google Scholar]
- 7. Donegan K, Matyac C, Seidler R, Porteous A. 1991. Evaluation of methods for sampling, recovery, and enumeration of bacteria applied to the phylloplane. Appl. Environ. Microbiol. 57:51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garsin DA, et al. 2001. A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. U. S. A. 98:10892–10897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gill HS, Rutherfurd KJ, Cross ML, Gopal PK. 2001. Enhancement of immunity in the elderly by dietary supplementation with the probiotic Bifidobacterium lactis HN019. Am. J. Clin. Nutr. 74:833–839 [DOI] [PubMed] [Google Scholar]
- 10. Hsu A-L, Murphy CT, Kenyon C. 2003. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300:1142–1145 [DOI] [PubMed] [Google Scholar]
- 11. Ikeda T, Yasui C, Hoshino K, Arikawa K, Nishikawa Y. 2007. Influence of lactic acid bacteria on longevity of Caenorhabditis elegans and host defense against Salmonella enterica serovar Enteritidis. Appl. Environ. Microbiol. 73:6404–6409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Irazoqui JE, Ng A, Xavier RJ, Ausubel FM. 2008. Role for β-catenin and HOX transcription factors in Caenorhabditis elegans and mammalian host epithelial-pathogen interactions. Proc. Natl. Acad. Sci. U. S. A. 105:17469–17474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Irazoqui JE, et al. 2010. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 6:e1000982 doi:10.1371/journal.ppat.1000982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Irazoqui JE, Urbach JM, Ausubel FM. 2010. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat. Rev. Immunol. 10:47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim DH, et al. 2002. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297:623–626 [DOI] [PubMed] [Google Scholar]
- 16. Kim Y, Oh S, Kim SH. 2009. Released exopolysaccharide (r-EPS) produced from probiotic bacteria reduce biofilm formation of enterohemorrhagic Escherichia coli O157:H7. Biochem. Biophys. Res. Commun. 379:324–329 [DOI] [PubMed] [Google Scholar]
- 17. Kim Y, Oh S, Yun HS, Kim SH. 2010. Cell-bound exopolysaccharide from probiotic bacteria induces autophagic cell death of tumour cells. Lett. Appl. Microbiol. 51:123–130 [DOI] [PubMed] [Google Scholar]
- 18. Konstantinov SR, et al. 2008. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc. Natl. Acad. Sci. U. S. A. 105:19474–19479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lebeer S, Vanderleyden J, De Keersmaecker SCJ. 2010. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat. Rev. Micro. 8:171–184 [DOI] [PubMed] [Google Scholar]
- 20. Lee J, Jayaraman A, Wood T. 2007. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 7:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee KZ, Kniazeva M, Han M, Pujol N, Ewbank JJ. 2010. The fatty acid synthase fasn-1 acts upstream of WNK and Ste20/GCK-VI kinases to modulate antimicrobial peptide expression in C. elegans epidermis. Virulence 1:113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liberati NT, et al. 2004. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc. Natl. Acad. Sci. U. S. A. 101:6593–6598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 24. Matsuzaki T, Chin J. 2000. Modulating immune responses with probiotic bacteria. Immunol. Cell Biol. 78:67–73 [DOI] [PubMed] [Google Scholar]
- 25. Medzhitov R. 2007. Recognition of microorganisms and activation of the immune response. Nature 449:819–826 [DOI] [PubMed] [Google Scholar]
- 26. Metchnikoff E. 1907. The prolongation of life: optimistic studies. Heinemann, London, United Kingdom [Google Scholar]
- 27. Moy TI, et al. 2006. Identification of novel antimicrobials using a live-animal infection model. Proc. Natl. Acad. Sci. U. S. A. 103:10414–10419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murphy CT, et al. 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424:277–283 [DOI] [PubMed] [Google Scholar]
- 29. Paulsen IT, et al. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074 [DOI] [PubMed] [Google Scholar]
- 30. Plantinga TS, et al. 2011. Differential Toll-like receptor recognition and induction of cytokine profile by Bifidobacterium breve and Lactobacillus strains of probiotics. Clin. Vaccine Immunol. 18:621–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Powell JR, Kim DH, Ausubel FM. 2009. The G protein-coupled receptor FSHR-1 is required for the Caenorhabditis elegans innate immune response. Proc. Natl. Acad. Sci. U. S. A. 106:2782–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pujol N, et al. 2001. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr. Biol. 11:809–821 [DOI] [PubMed] [Google Scholar]
- 33. Pukkila-Worley R, Ausubel FM, Mylonakis E. 2011. C. albicans infection of Caenorhabditis elegans induces antifungal immune defenses. PLoS Pathog. 7:e1002074 doi:10.1371/journal.ppat.1002074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reid G, et al. 2011. Microbiota restoration: natural and supplemented recovery of human microbial communities. Nat. Rev. Microbiol. 9:27–38 [DOI] [PubMed] [Google Scholar]
- 35. Shivers RP, et al. 2010. Phosphorylation of the conserved transcription factor ATF-7 by PMK-1 p38 MAPK regulates innate immunity in Caenorhabditis elegans. PLoS Genet. 6:e1000892 doi:10.1371/journal.pgen.1000892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sifri CD, Begun J, Ausubel FM. 2005. The worm has turned—microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 13:119–127 [DOI] [PubMed] [Google Scholar]
- 37. So S, Tokumaru T, Miyahara K, Ohshima Y. 2011. Control of lifespan by food bacteria, nutrient limitation and pathogenicity of food in C. elegans. Mech. Ageing Dev. 132:210–212 [DOI] [PubMed] [Google Scholar]
- 38. Tan M-W, Mahajan-Miklos S, Ausubel FM. 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 96:715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tenor JL, Aballay A. 2008. A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO Rep. 9:103–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thomas CM, Versalovic J. 2010. Probiotics-host communication: modulation of signaling pathways in the intestine. Gut Microbes 1:148–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Troemel ER, et al. 2006. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2:e183 doi:10.1371/journal.pgen.0020183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vance RE, Isberg RR, Portnoy DA. 2009. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe 6:10–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang C, et al. 2011. Use of Caenorhabditis elegans for preselecting Lactobacillus isolates to control Salmonella Typhimurium. J. Food Prot. 74:86–93 [DOI] [PubMed] [Google Scholar]
- 44. Wang X, Wang X, Li L, Wang D. 2010. Lifespan extension in Caenorhabditis elegans by DMSO is dependent on sir-2.1 and daf-16. Biochem. Biophys. Res. Commun. 400:613–618 [DOI] [PubMed] [Google Scholar]
- 45. Wells JM, Mercenier A. 2008. Mucosal delivery of therapeutic and prophylactic molecules using lactic acid bacteria. Nat. Rev. Microbiol. 6:349–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wong D, Bazopoulou D, Pujol N, Tavernarakis N, Ewbank J. 2007. Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol. 8:R194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Y, Lu H, Bargmann CI. 2005. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438:179–184 [DOI] [PubMed] [Google Scholar]
- 48. Zugasti O, Ewbank JJ. 2009. Neuroimmune regulation of antimicrobial peptide expression by a noncanonical TGF-β signaling pathway in Caenorhabditis elegans epidermis. Nat. Immunol. 10:249–256 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.