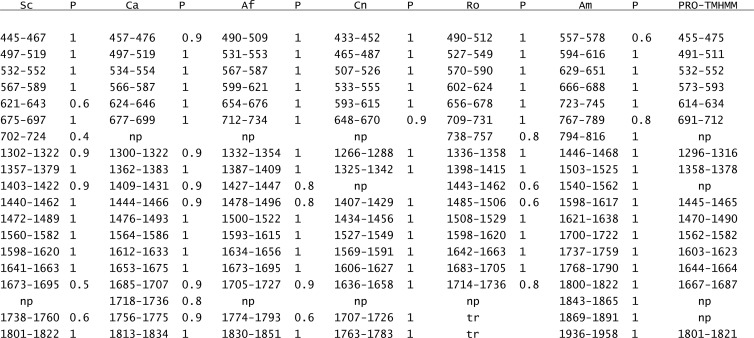
Fig 1.
TMHMM-predicted TMH locations (residue number) and probabilities (P) for Fks1 from S. cerevisiae (Sc), C. albicans (Ca), A. fumigatus (Af), C. neoformans (Cn), R. oryzae (Ro), and A. macrogynus (Am). The PRO-TMHMM prediction was generated using S. cerevisiae Fks1 as a query. TMH on the same line are considered to be equivalent based on overlap in the Fks1 ClustalW2 alignment (see Fig. S1 in the supplemental material). np, not predicted; tr, truncated C terminus.