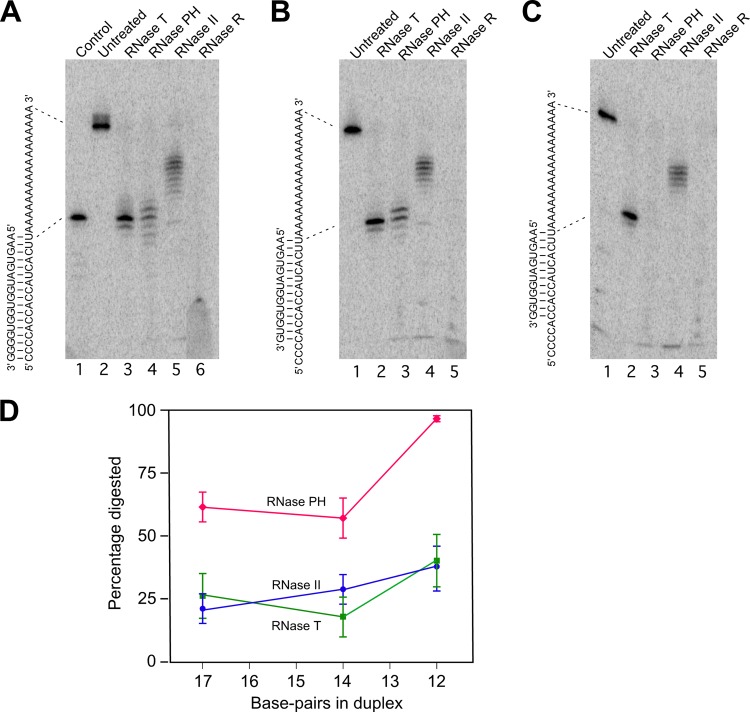
Fig 3.
Digestion of duplex RNA by exonucleases. (A to C) Three partial duplex substrates—duplex I, duplex II, and duplex III—were prepared by annealing a 5′-labeled 34-mer RNA with 17-, 14-, and 12-nt complementary oligonucleotides. Each substrate was untreated or treated with the indicated RNase, and the extent of substrate digestion was assessed following gel electrophoresis. The sequence of each duplex is shown on the left of each panel. The migration of the undigested labeled RNA and a product derived by the removal of the 3′ single-stranded residues from the labeled RNA are indicated by dotted lines. (A) Digestion of duplex I. A 17-nt control RNA that corresponds to the part of the labeled RNA that is base-paired in duplex I was also run on the gel (lane 1). (B) Digestion of duplex II. (C) Digestion of duplex III. (D) For each substrate, phosphorimaging of dried gels was performed to quantify the extent of digestion through the duplex region with each enzyme. The mean extent of digestion and standard errors by each enzyme for the different duplexes, derived from five experiments, is indicated. Blue circles, digestion by RNase II; green squares, digestion by RNase T; red diamonds, digestion by RNase PH.