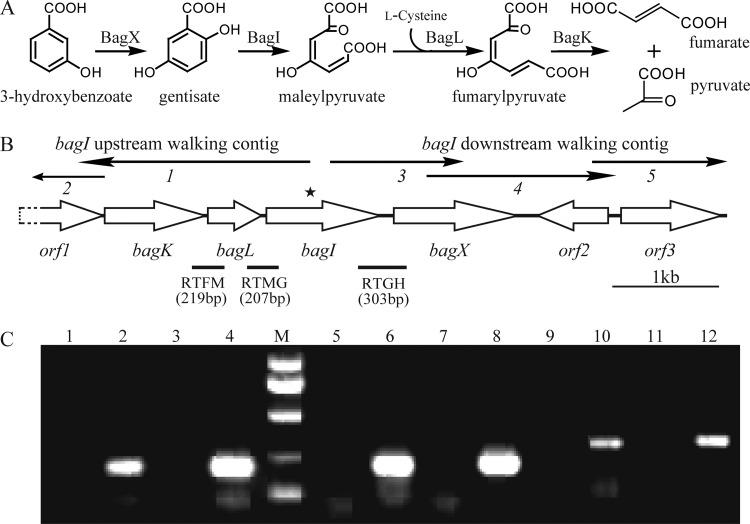
Fig 1.
(A) Proposed pathway for the 3-hydroxybenzoate catabolism via gentisate in Paenibacillus sp. strain NyZ101, together with the catabolic reactions catalyzed by the bag gene products in vivo. (B) Organization of the bag gene cluster of Paenibacillus sp. strain NyZ101. The arrows indicate the size and direction of transcription of each gene or ORF. The star above bagI indicates the region where the genome walking started in both directions. The thin arrows above the gene cluster represent the fragments obtained by genome-walking reactions. The locations of the primer sets RTFM, RTMG, and RTGH and the amplified DNA fragments for RT-PCR are indicated below. (C) Analysis of bagILKX transcription by RT-PCR. Reactions performed without RT were used as negative controls, as shown in lanes 1, 3, 5, 7, 9, and 11. Lane M, molecular size markers at 1,000, 750, 500, 250, and 100 bp. Lanes 2, 6, and 10 (template from succinate-grown strain NyZ101) and lanes 4, 8, and 12 (template from succinate-grown strain NyZ101 induced by 3HBA) show products amplified using the RTFM, RTMG, and RTGH primer sets, respectively, with products of RT.