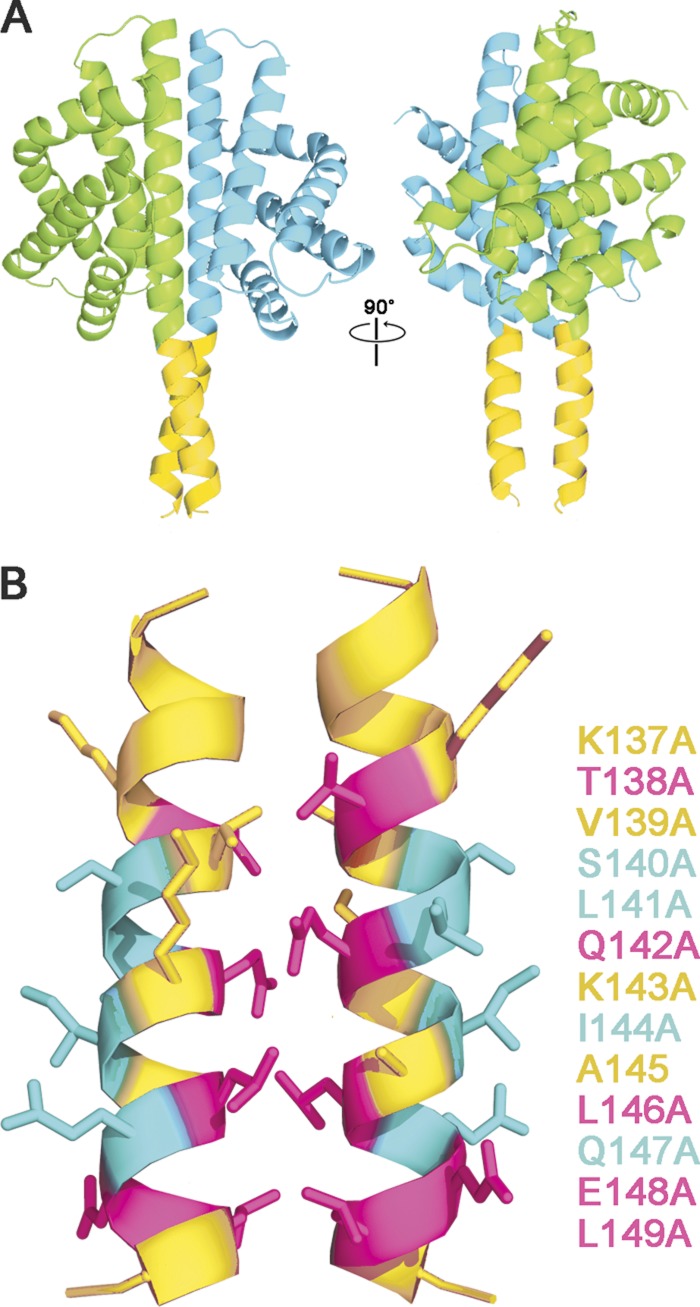
Fig 6.
Computational model of paired RsbRA linker helices, derived by extending the H helices of the nonheme globin crystal structure (PDB code 2BNL, chains A and B, cyan and green ribbons) to include the linker helices (gold ribbons, RsbRA residues 137 to 150; see Materials and Methods). (A) Two views of the nonheme globin dimer showing crossover of the linker helices at Q142 (left) and a 90° rotated view looking into the putative interaction surfaces of the paired helices (right). (B) Details of the interaction surfaces, indicating the effects of alanine substitution at positions 137 to 149: substitutions that enhance steady-state output of the stressosome (magenta), those that diminish output (cyan), and those with no significant effect (gold). This correspondence of helical periodicity with mutational phenotype is consistent with the notion that each linker helix interacts with two different binding partners: its counterpart helix via the highly conserved interface residues and an unknown partner via the nonconserved residues on the opposite face.