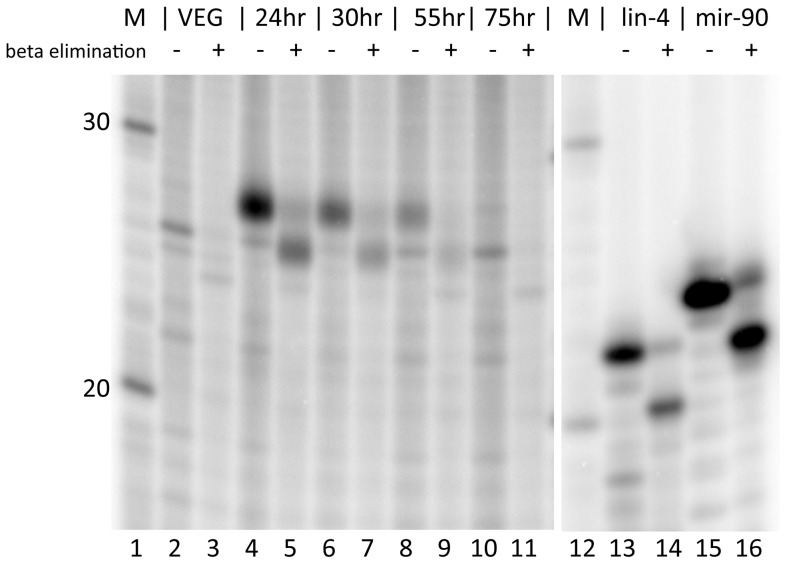
Figure 2. The mating-specific 27 nt RNAs in Oxytricha trifallax are not modified at their 3′ ends.
RNAs were tested with the beta elimination assay in order to determine if there is a modification at the 3′ end. RNAs tested are 32P 5′ end labeled total Oxytricha trifallax RNA (lanes 2–11) or 5′ end labeled synthetic positive control RNAs containing the sequence of the C. elegans lin-4 (21 nt) or mir-90 (23 nt) microRNAs which were subsequently mixed with 1.5 µg of unlabeled Oxytricha total RNA prior to beta elimination (lanes 13–16). Control untreated samples (-) or samples subjected to the beta elimination reaction (+) are indicated. RNAs were isolated from vegetative ALXC2 (lanes 2 and 3) or from a mating between ALXC2 and ALXC9 that were harvested at the indicated timepoint after mixing (lanes 4–11). Beta elimination will remove the terminal ribose if both a free 2′ OH and 3′ OH are present, and will leave a 3′ cyclic phosphate. This loss of a nucleotide and the presence of the extra phosphate group will result in the treated RNAs running almost two bases faster than their untreated counterparts.