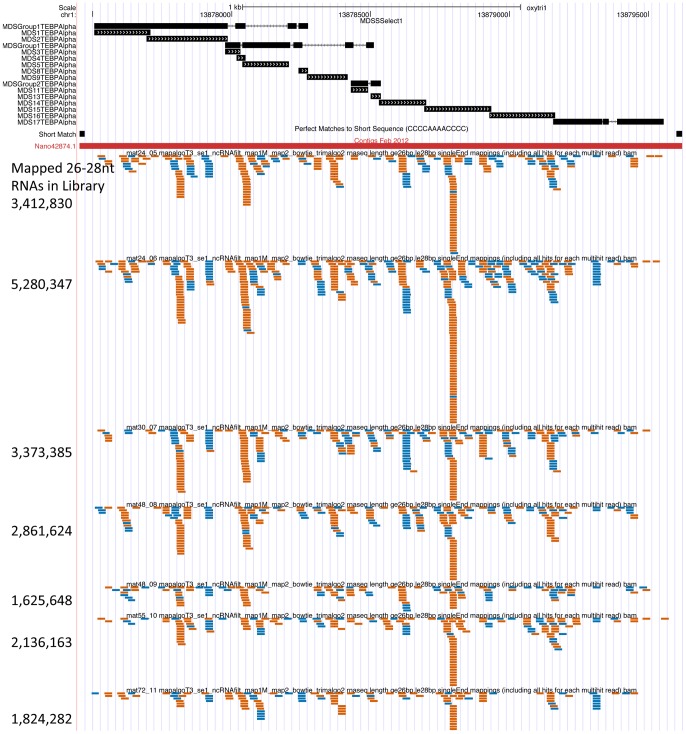
Figure 6. 27 nt RNA reads from different mating libraries align to both strands of a nanochromosome.
This screen shot from the Oxytricha trifallax macronuclear genome build of the UCSC Genome Browser shows the macronuclear nanochromosome corresponding to the alpha telomere binding protein gene indicated as a red bar (Nano42874.1) with the telomere sequences at the beginning and end of the nanochromosome noted as small black boxes in the track above that. The top track shows the locations of groups of MDS sequences or individual MDSs, as aligned using BLAT of the micronuclear sequence against the macronuclear genome. The alignments of 26–28 nt small RNAs from 7 different mating small RNA libraries (mat24_05, mat24_06, mat30_07, mat48_08, mat48_09, mat55_10 and mat72_11 - see Table 1) are shown with alignments to the plus strand of the nanochromosome indicated in blue and alignments to the minus strand of the nanochromosome indicated in orange. Numbers at the left indicate the total number of 26, 27 and 28 nt RNA sequences from each of the seven libraries that mapped to the macronuclear genome.