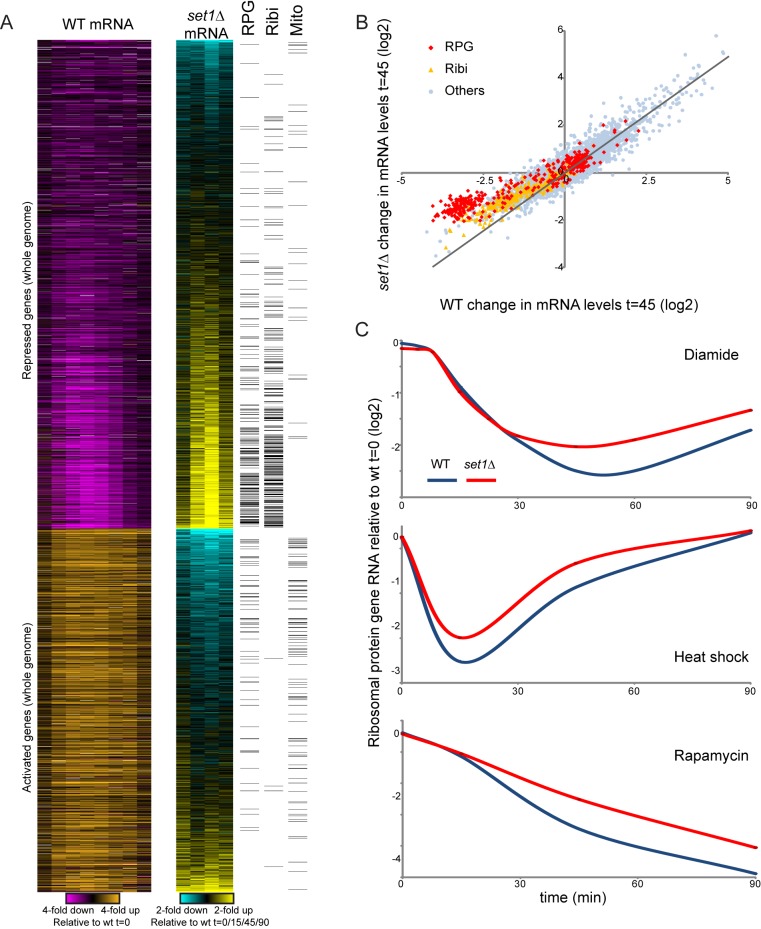
Figure 5. Set1 is predominantly a repressor during diamide stress.
(A) Whole genome analysis of set1Δ effects on diamide stress response. Left panel: gene expression data from Gasch et al. [21] for all genes induced or repressed at least 1.8-fold. Middle panel: effect of set1Δ on diamide stress, for t = 0, 15, 45, or 90 min, using whole-genome microarray data. Genes are grouped by repressed/activated, then subsequently sorted by the average set1Δ effect on gene expression. Right panel: ribosomal protein (RPG) or ribosomal biogenesis (Ribi) GO annotations for individual genes are indicated as black bars. The majority of genes that repress poorly in set1Δ mutants are involved in cytoplasmic ribosome biosynthesis. (B) Set1 functions primarily as a repressor. Scatterplot of change in mRNA abundance in wild-type (x-axis) or set1Δ (y-axis) yeast at diamide t = 45 min. There is overall excellent correlation between the two datasets except for blunted repression of ribosomal protein genes and ribosomal biogenesis genes, as indicated. (C) Set1 affects ribosomal gene repression in response to multiple distinct environmental conditions. Wild type and set1Δ yeast were subjected to time courses of diamide stress, 37°C heat shock, or 0.2 µg/mL rapamycin. RNA abundance was measured by nCounter, and normalized ribosomal protein gene RNA abundances are averaged and shown as log2 abundance. Note that for all three stresses, Set1 is required for full RPG repression.