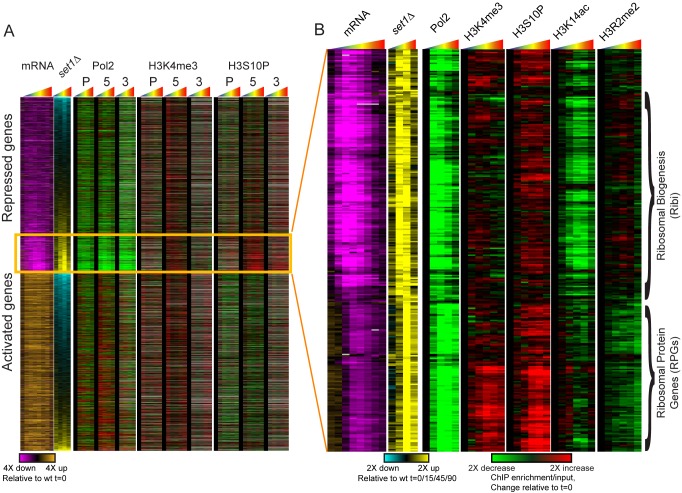
Figure 6. Specific chromatin changes occur at RPG and Ribi genes during repression.
(A) Whole genome mRNA [21], set1Δ effects on mRNA (this study), Pol2 mapping [26], and tiling microarray data for H3K4me3 and H3S10P (this study) are shown for all genes sorted as in Figure 5A. All four datasets represent a time course of diamide response, as indicated by rainbow triangles above each box. (B) RPG and Ribi genes exhibit distinct chromatin changes during diamide stress. Diamide-repressed genes whose repression is diminished in set1Δ mutants were clustered according to their associated changes in chromatin marks. Tiling microarray data are shown only for 5′CDS probes for each mark. A clear separation can be observed between RPGs, which exhibit increased 5′ H3K4me3 and decreased 5′ H3R2me2, and Ribi genes, which exhibit decreased 5′ H3K14ac.