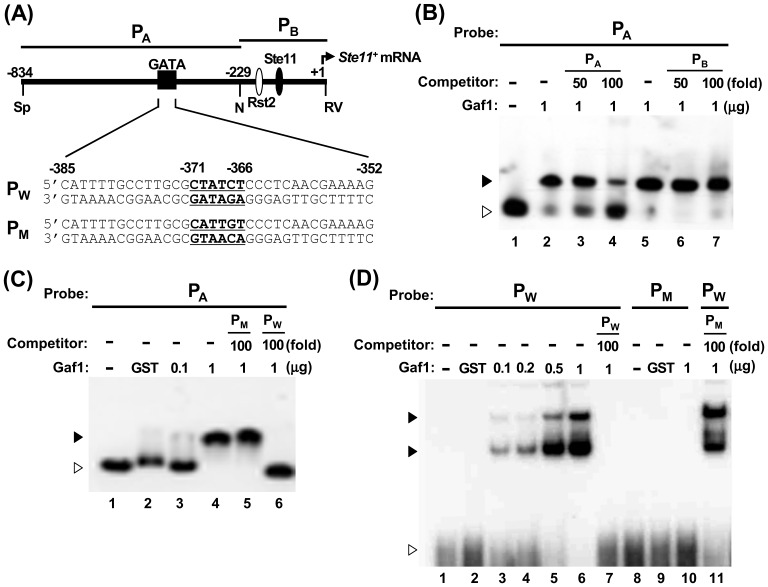
Figure 6. Gaf1 protein specifically binds to the GATA binding motif of ste11+ promoter in vitro.
(A) Schematic diagram of the promoter region of ste11+. The open- and closed-ovals represent binding sites for Rst2 (UASst) and Ste11 (TR1 and TR2), respectively. The major transcription start site (position number +1) is designated by a hinged arrow. The regions contained in the upstream probe PA (−834∼−225) and the downstream probe PB (−231∼+10) are shown as bars. The dark square represents the GATA motif spanning from −385 to −352 for which wild-type (PW) and mutant (PM) double-stranded oligonucleotide probes were designed. Restriction sites: Sp, SphI; N, NdeI; RV, EcoRV. (B) Search for Gaf1-binding region in ste11+ promoter by EMSA. GST-Gaf1 protein (1 µg) was incubated with labeled PA probe containing the 0.6-kb upstream region of ste11+ promoter in the absence (lanes 2 and 5) or presence of excess cold competitor, PA (lane 3, 50-fold; lane 4, 100-fold) or PB (lane 6, 50-fold; lane 7, 100-fold). (C) Analysis of Gaf1 binding to the GATA motif of ste11+ promoter by EMSA. GST-Gaf1 protein (lane 3, 0.1 µg; lanes 4–6, 1 µg) was incubated with labeled PA probe in the absence (lanes 3 and 4) or presence of 100-fold excess of cold competitor PM (lanes 5) or PW (lanes 6). (D) Mutational dissection of the Gaf1-binding GATA motif of ste11+ promoter by EMSA. Varying amounts of GST-Gaf1 protein (lane 3, 0.1 µg; lane 4, 0.2 µg; lane 5, 0.5 µg; lanes 6, 7, 11, and 12, 1 µg) were incubated with labeled PW oligonucleotide probe in the absence (lanes 3 and 4) or presence of 100-fold excess of cold competitor PM (lanes 5) or PW (lanes 6). Mock reaction mixtures without GST-Gaf1 or with GST protein were used as negative controls in (B)–(D). The closed and open arrow heads in (B)–(D) represent shifted bands and free probes, respectively.