Table 3. List of the Ste11 target genes up-regulated by gaf1Δ mutation.
| Expression ratioa | ||||||
| Subgroup | Systematic No. | Gene | Description (GeneDB) |
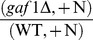
|
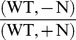
|
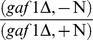
|
| (G)b | SPAC27D7.03c | mei2 + | RNA-binding protein involved in meiosis | 4.07 | 43.74 | 9.30 |
| SPAC32A11.01 | mug8 + | conserved fungal protein | 2.27 | 5.82 | 4.25 | |
| SPAC31G5.09c | spk1 + | MAP kinase | 1.95 | 36.46 | 23.90 | |
| SPBC32C12.02 | ste11 + | transcription factor | 1.76 | 5.87 | 3.10 | |
| SPBC359.06 | mug14 + | adducing | 1.73 | 82.76 | 45.98 | |
| SPAC11E3.06 | map1 + | MADS-box transcription factor | 1.72 | 11.77 | 6.14 | |
| SPBC19C2.04c | ubp11 + | ubiquitin C-terminal hydrolase | 1.72 | 5.07 | 3.84 | |
| SPBC24C6.06 | gpa1 + | G-protein alpha subunit | 1.70 | 11.54 | 7.32 | |
| SPAC1565.04c | ste4 + | adaptor protein | 1.69 | 10.87 | 5.41 | |
| SPCC162.10 | ppk33 + | serine/threonine protein kinase | 1.53 | 46.35 | 30.03 | |
| (D+E)c | SPAC1F5.09c | shk2 + | PAK-related kinase | 1.27 | 20.72 | 53.30 |
| SPCC1442.01 | ste6 + | guanyl-nucleotide exchange factor | 0.98 | 7.40 | 15.76 | |
| SPAC1093.06c | dhc1 + | dynein heavy chain | 0.30 | 1.47 | 15.66 | |
| SPAC23E2.03c | ste7 + | meiotic suppressor protein | 1.19 | 11.05 | 15.28 | |
| SPBC19C2.05 | ran1 + | serine/threonine protein kinase | 1.18 | 5.38 | 6.88 | |
| SPCC1393.07c | mug4 + | sequence orphan | 1.17 | 1.97 | 5.57 | |
| SPAC31G5.07 | conjugation protein (predicted) | 0.89 | 1.52 | 4.45 | ||
| SPBC354.08c | DUF221 family protein | 0.36 | 0.92 | 4.07 | ||
| SPBC2D10.06 | rep1 + | MBF transcription factor complex subunit | 0.92 | 2.00 | 3.36 | |
| SPAC13C5.03 | tht1 + | nuclear membrane protein involved in karyogamy | 1.04 | 0.62 | 3.01 | |
| SPAC1F5.08c | yam8 + | calcium transport protein | 1.07 | 1.55 | 2.65 | |
| SPAC27E2.07 | pvg2 + | galactose residue biosynthesis protein | 1.21 | 1.59 | 2.48 | |
| SPAPB2B4.03 | cig2 + | cyclin | 0.80 | 1.09 | 2.39 | |
| SPAC31G5.10 | eta2 + | Myb family protein | 1.04 | 1.41 | 2.22 | |
| SPBP4H10.11c | lcf2 + | long-chain-fatty-acid-CoA ligase | 0.75 | 1.02 | 2.08 | |
Relative expression of the genes was measured by microarray assay in the nitrogen-starved (−N) and unstarved (+N) cells of wild-type (ED668) and gaf1Δ (KL216) strains. Unstarved (+N) cells were prepared from the mid-log phase cultures in EMM2. Nitrogen-starved (−N) cells were prepared by shifting the mid-log phase cells to EMM-N for 4 h as described in Materials and Methods.
Genes up-regulated by gaf1Δ mutation under both nitrogen-starved and unstarved conditions (Subgroup G, p<0.05).
Genes up-regulated by gaf1Δ mutation only under nitrogen starved conditions (Subgroup D+E, p<0.05).
