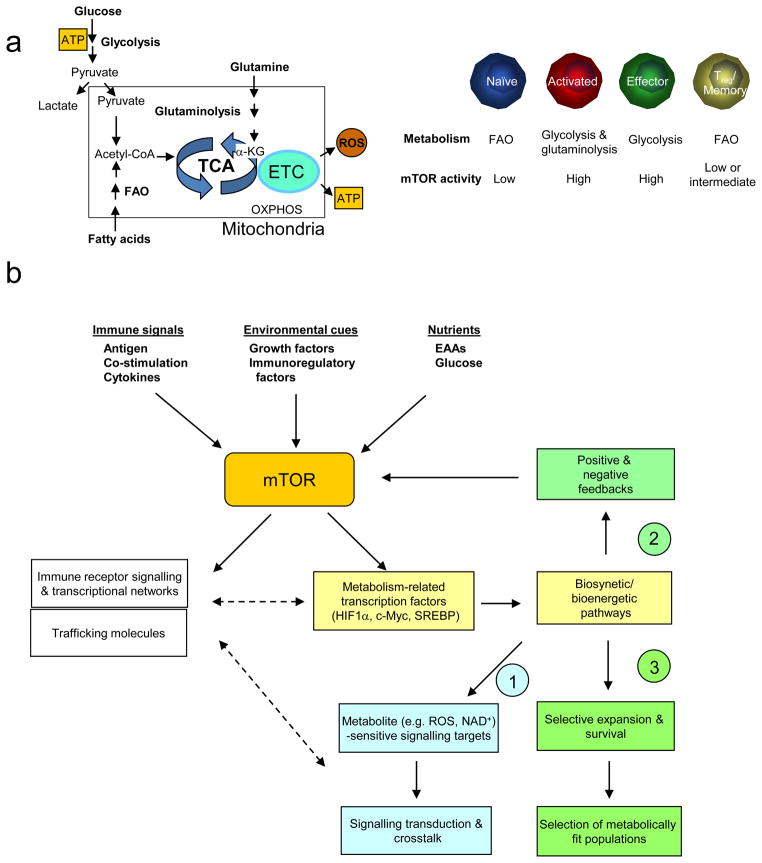
Figure 3. mTOR in T cell metabolism.
(a) Metabolic programmes in T cells (left) and mTOR activity in T cell subsets (right). Only the prominent metabolic programs are presented. FAO, fatty acid oxidation; TCA, tricarboxylic-acid cycle; ETC, electron-transport chain; ROS, reactive oxygen species; OXPHOS, oxidative phosphorylation.
(b) Proposed mechanisms of mTOR-mediated metabolic pathways in T cell differentiation. An important upstream signal to activate mTOR is nutrients, in particular essential amino acids (EAAs) whose levels are actively controlled by dendritic cells (DCs). Once activated, mTOR serves as a platform to engage several downstream effector pathways including immune receptor signalling, metabolic programme and migratory activity. mTOR promotes metabolism via activating a gene expression programme consisting of metabolic gene targets of transcription factors hypoxia-inducible factor 1α (HIF1α), c-Myc and SREBP, which in turn impinge upon biosynthetic and bioenergetic pathways. Three potential downstream mechanisms are proposed to explain how these metabolic pathways regulate T cell differentiation, including (1) signal crosstalk; (2) feedback control; (3) selective expansion and survival. Among metabolites with signalling activities, reactive oxygen species (ROS) oxidize the catalytic cysteine residues of phosphatases to cause their inactivation, whereas NAD+ is required for the activity of sirtuin family deacetylases.