Figure 2. Evidence for THF riboswitch function.
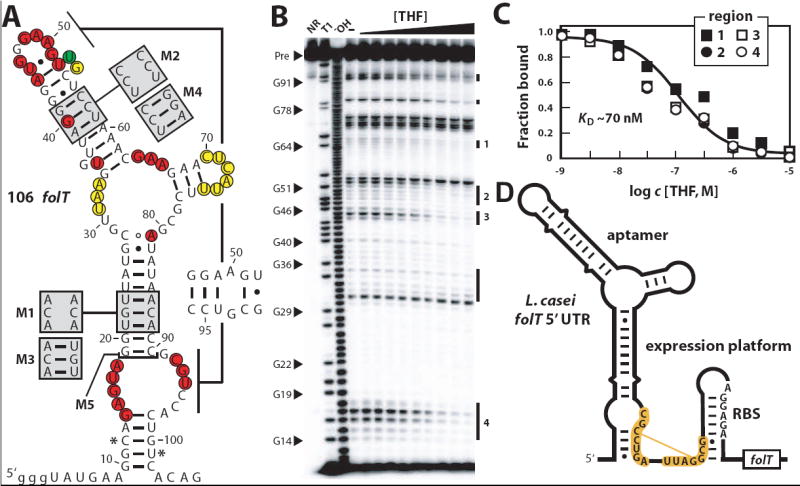
(A) Sequence and predicted secondary structure of the 106 folT RNA from A. metalliredigens. Nucleotides shaded yellow, red or green identify internucleotide linkages that undergo constant, decreasing or increasing scission when THF is added to an in-line probing assay (see gel image in B). Asterisks identify the 5′ and 3′ boundaries of the in-line probing annotations, which are restricted due to limitations of product separation by polyacrylamide gel electrophoresis. Shaded boxes designate sequence changes present in mutants M1 through M4 as indicated. M5 construct is truncated at the bracket. Lowercase letters identify nucleotides added to the construct to facilitate efficient in vitro transcription. (B) In-line probing analysis of the A. metalliredigens 106 folT RNA. NR, T1 and −OH designate precursor (Pre) RNAs subjected to no reaction, partial digestion with RNase T1 (cleaves after G residues), and partial digest under alkaline conditions (cleaves at every position). Other lanes include Pre RNAs subjected to in-line probing reactions without THF (–) or with increasing concentrations of THF (1 nM to 10 μM). Vertical lines identify cleavage products whose yields are altered by THF addition, and numbered regions were quantified and used to estimate KD values. (C) Plot of the fraction of RNAs bound to ligand versus the logarithm of the concentration of THF. The solid line represents a theoretical binding curve for a one-to-one interaction with a KD of 70 nM. Fraction bound values were generated by quantifying band intensities from the regions identified in B. (D) A possible THF riboswitch expression platform is evident in the L. casei folT mRNA. The nucleotides shaded in orange can form an alternative base pairing interaction in the absence of THF that may interfere with the formation of the putative anti-RBS hairpin.
