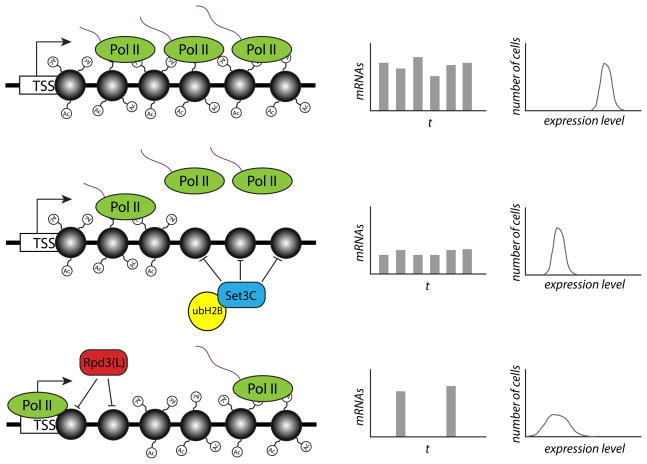
Figure 1. The HDACs Set3C and Rpd3(L) act at distinct steps of transcription to modulate either transcriptional burst size or burst frequency.
The figure illustrates the effect of histone deacetylation by either Set3C or Rpd3(L) on the expression and noise levels of a target gene. In the top row, RNA polymerase II (Pol II) transcribes RNA (purple strands) from a gene with heavily acetylated nucleosomes (grey circles), leading to high-frequency transcriptional bursts of relatively large size. When Set3C and ubH2B act to deacetylate nucleosomes in the body of the gene (middle row), it causes a decrease in Pol II processivity and an increased likelihood of abortive transcription. This affects the average size of each transcriptional burst but not the frequency, resulting in a decrease in mean expression levels but the same level of noise. Rpd3(L), by contrast, acts to deacetylate nucleosomes just downstream of the transcription start site (TSS), leading to a reduced frequency of transcriptional initiation and bursts (bottom row). This lower burst frequency results in both lower mean expression levels and increased noise levels, as seen by the increased width of the expression distribution.