FIGURE 2. Early increase of TGF-β mRNA transcript after intravenous donor apoptotic cell infusion simultaneously with bone marrow graft.
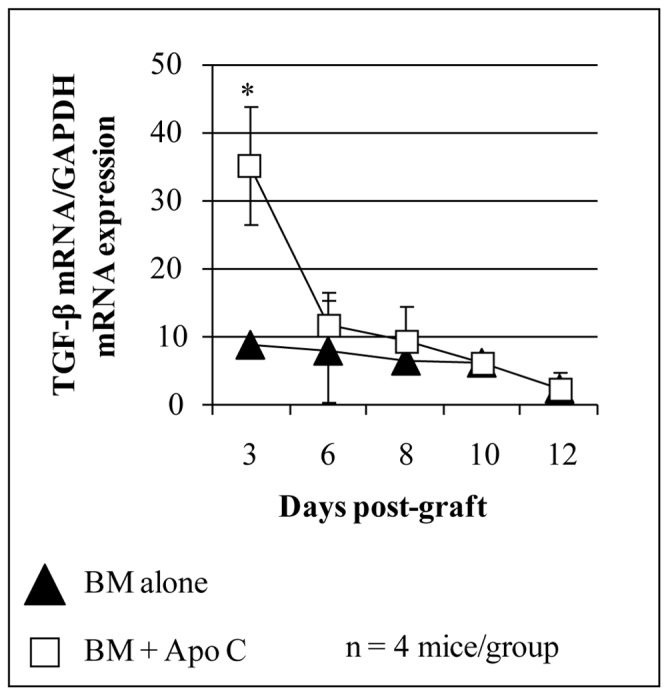
Total RNA was extracted from spleen of BALB/c mice grafted with 106 FVB BM (black triangles) or with 5x106 FVB apoptotic splenocytes (Apo C) plus 106 FVB BM (open square) using RNA extraction kit (Qiagen, Valencia, CA) and reverse transcribed using random hexamers and M-MLV reverse transcriptase (Life Technologies, Rockville, MD) to use as template for quantitative real time RT-PCR (QRT-PCR). QRT-PCR reactions were performed as described (18). Primers and dual labeled fluorescent probes were designed using Primer Express® software (Applied Biosystems, Forster City, CA) to be specific for RNA but not for genomic DNA. QRT-PCR primer pairs and related probes were as follow (sense, antisense and probe respectively). TGF-β : 5′-GCTCTTGTGACAGCAAAGATAACAA-3′, 5′-GGTCGCCCCGACGTTT-3′ and 5′-FAM-CACGTGGAAATCAACGGGATCAGCC-TAMRA-3′; Primer pairs for GAPDH (used as endogenous reference)were already described (18). The relative quantity of each unknown sample was determined automatically with the iCycler iQ® software (Bio-Rad Laboratories, Marnes-la-Coquette, France), using the threshold cycle (Ct). Data were expressed as normalized TGF-β1 expression, which was obtained by dividing the relative quantity of TGF-β1 mRNA for each sample by the relative quantity of GAPDH mRNA of the same sample. *P<0.05.
