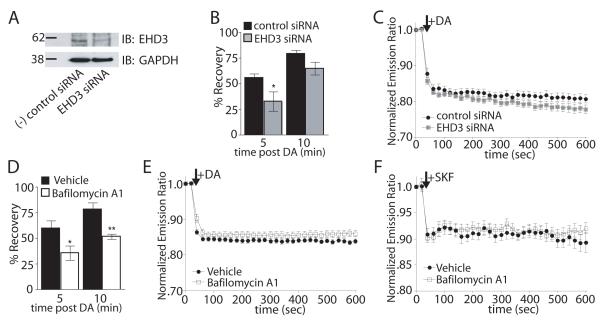
Figure 7. Inhibiting recycling does not affect acute D1 receptor-mediated cAMP accumulation.
(A) Representative (n = 3) immunoblot analysis of siRNA mediated knockdown of EHD3 in FD1R-expressing HEK293 cells with GAPDH loading control. (B) Recovery of surface FD1Rs measured by fluorescence flow cytometry. Recovery of surface FD1R fluorescence after 5 min DA incubation and 5 or 10 min after agonist washout was measured in cells transfected with siRNA targeting EHD3 (gray) or non-silencing control siRNA (black). %Recovery = [(Surface fluorescence after DA washout - average surface fluorescence after 5 min. DA incubation)/(average initial surface fluorescence - average surface fluorescence after 5 min. DA incubation)] × 100. Data = mean %Recovery +/− SEM, n = 3, 10,000 cells/well, each time point in triplicate. (C) Change in normalized Epac1-cAMPs FRET in response to 10μM DA in cells transfected with siRNA against EHD3 (grey squares) or non-silencing control siRNA (filled circles). Data = mean normalized FRET emission ratio at each time point +/− SEM, n = 20-28 cells per group. (D) Recovery of FD1Rs in the presence of 500nM Bafilomycin (white bars) or vehicle (black bars) as measured by fluorescence flow cytometry. Data = mean %Recovery +/− SEM, n = 3, 10,000 cells/well, each time point in triplicate. (E) Change in normalized Epac1-cAMPs FRET in response to 10μM DA in cells pretreated with Bafilomycin (open squares) or vehicle control (filled circles). Data = mean normalized FRET emission ratio at each time point +/− SEM, n = 29-30 cells per group. (F) Change in normalized Epac1-cAMPs FRET in response to 1μM SKF81927 in FD1 expressing striatal neurons pretreated with bafilomycin (open squares) or vehicle control (filled circles). Data = mean normalized FRET emission ratio at each time point +/− SEM, n = 11-14 neurons per group. (*p<0.05, **p<0.01, 2-tailed unpaired t-test).