Figure 8.
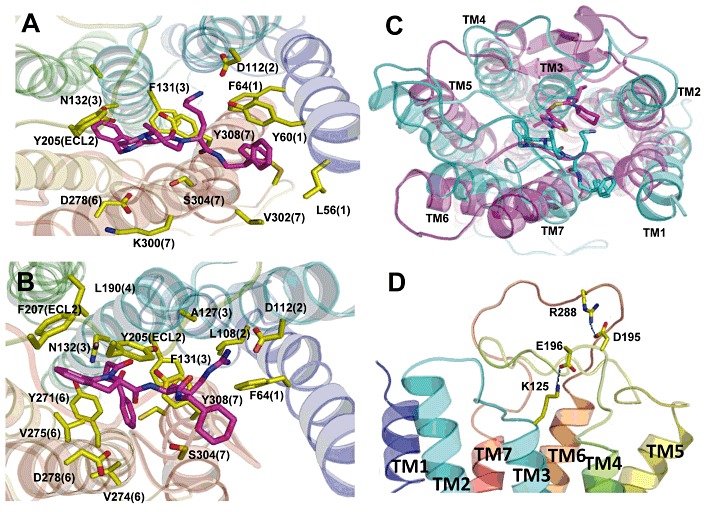
Ab initio modelling of CXCR3. (A) and (B) Top views of a model of human CXCR3 predicted using MembStruk showing the small molecule agonists Cp#1 (A) and Cp#3 (B) residing in a binding site predicted using Glide XP. In both models, D112 acts as a counter-ion for the basic moiety of either compound, with a predicted cluster of predominantly hydrophobic residues interacting with the ligand in the binding site. Numbers in parenthesis refer to the TM helix (1–7) in which the residue resides. (C) Comparison of the binding site of the compound called ‘1t’ in the CXCR4 crystal structure (pink) to the predicted binding site of Cp#3 in CXCR3 (cyan) shows that the two compounds bind in similar locations in their respective receptors. (D) The two predicted salt bridges within the extracellular domains of CXCR3 between K125 (ECL1) and D196 (ECL2) and between R288 (ECL3) and D195 (ECL2).
