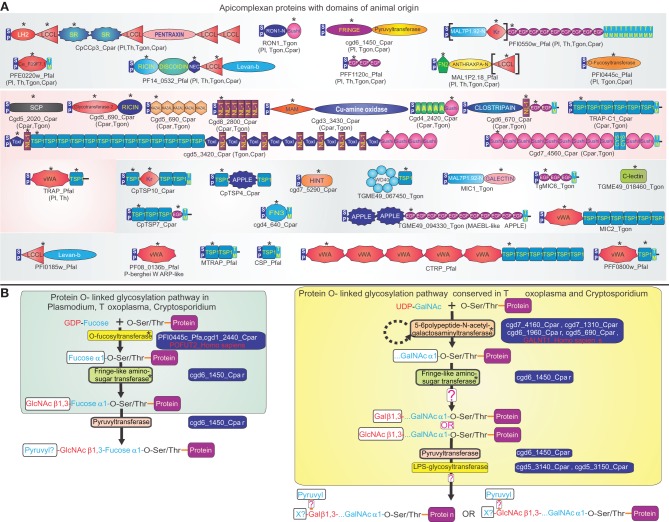
Figure 1.
Animal domains and animal-type O-glycosylation systems in apicomplexa. (A) Domain architectures of apicomplexan proteins containing adhesion domains of animal origin. Proteins are labeled by their gene names/common names and species abbreviation separated by an underscore, and are grouped based on their conservation in apicomplexans. If a domain architecture is present in more than one distinct apicomplexan lineage, the additional lineages are shown in brackets. Domains of animal origin are marked with an asterisk above the domain. If a domain is present in multiple copies in a protein, only one (the first) instance of it is labeled with an asterisk. Domains not present in all orthologs of a protein are enclosed in square brackets. Standard abbreviations are used for domains. Species abbreviations are as follows: Cpar: Cryptosporidium parvum, Pl: Plasmodium, Pfal: Plasmodium falciparum, Th: Theileria, Tgon: Toxoplasma gondii. (B) Protein O-linked glycosylation pathways of animal provenance in apicomplexans. Gene names of enzymes involved in these pathways are shown to the right of the enzyme, along with examples of orthologous proteins from animals. The reconstructed oligosaccharide chain is represented using abbreviations for various sugars and functional groups. Speculative parts are marked with a “?”. GalNAc: N-acetylgalactosamine, GlcNAc: N-acetylglucosamine, X? indicates an uncharacterized sugar added by the LPS glycosyltransferase. Enzymes of animal origin are marked with an asterisk. Species abbreviations are as in (A).