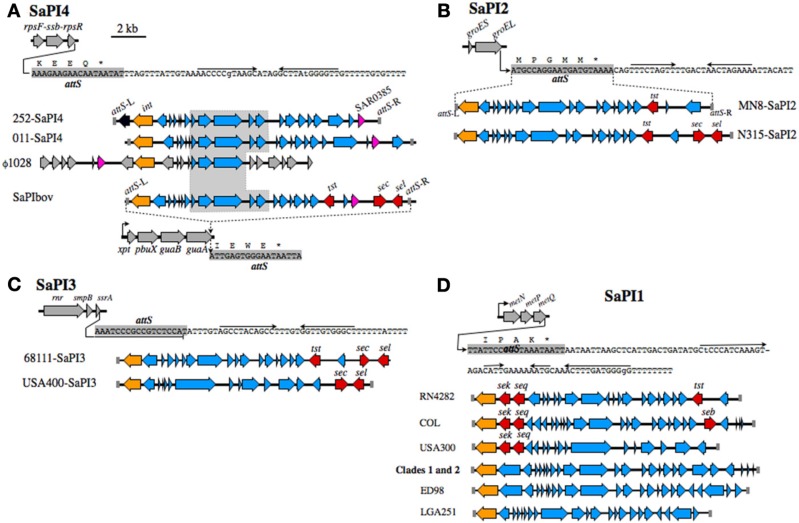
Figure 2.
SaPI structures and insertion sites. (A) SaPI4, with an illustration of the rpsF-ssb-rpsR operon, and nucleotide sequence extending from the 3′-end of rpsR. The attS of SaPI4 shaded gray, overlaps the 3′-end of the rpsR open reading frame, and letters above the sequence correspond to the C-terminus of RpsR protein. Arrows above the nucleotide sequence indicate inverted repeats, likely comprising a transcriptional terminator. Illustrations below the sequence compare SaPI4 of S. aureus MRSA 252 with that of ovine adapted strain 011. The duplicated left and right attS sequences are shaded gray, int genes are colored orange, while SAR0385 and orthologous genes, encoding a putative secreted protein, are magenta. A central outlined and shaded segment of SaPI4 is highly conserved in S. aureus phage ϕ1028, and SaPIbov of bovine adapted S. aureus ET3-1. Genes in SaPIbov encoding superantigen toxins are shaded red. The int of SaPIbov is shaded orange to emphasize function, although each SaPI family has a distinct int and attS. Structures of SaPI2 (B), SaPI3 (C), SaPI1 (D), and their genomic insertion sites. Features are labeled as in A. SaPI2 of ST30spa33 strain MN8 (B), is compared to SaPI2 of a non-related MRSA strain N315. SaPI3 (C) is not present in CC30. Structures shown are from strain USA400 [Baba et al. (2002)], which is a CA-MRSA, and strain 68111, which is a triple locus MLST variant of ST30 [Li et al. (2011)]. A clonal complex is comprised of isolates that differ from the ancestral sequence type (ST) at no more than two of seven MLST alleles. Therefore, S. aureus 68111 is distantly related to ST30, but is not within CC30. (D) shows SaPI1 structures from different strains, in comparison to SaPI1 in Clade 1 PT80/81 strains, and Clade 2 ST30spa19 CA-MRSA.