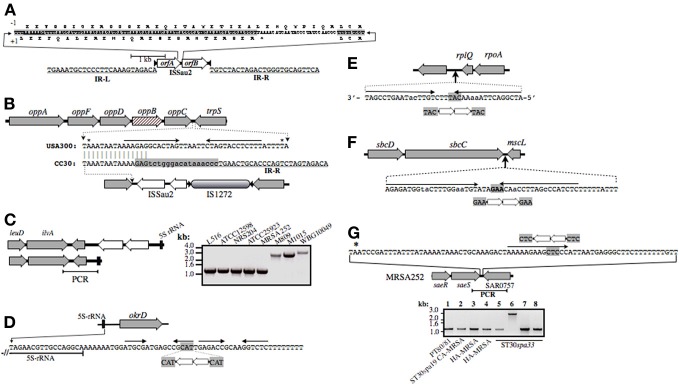
Figure 3.
Illustration of ISSau2 (A) and its insertion sites (B–G) in CC30. ISSau2 is comprised of orfA and orfB (A), flanked by 39 nt inverted repeats IR-L and IR-R, with terminal 5′-TG and 3′-CA dinucleotides (A). The sequence above the illustration spans the 3′-end of orfA and 5′-end of orfB. The +1 translation of orfA, terminating at a stop codon, is shown below the sequence. Above the sequence is a translation that would result from a −1 frame-shift within AAAAAAG. The −1 translation from this point onwards continues throug to the end of orfB, and would produce a single 1569 nt trans-frame protein. (B) Illustration of oppAFBDC and trpS genome segment of non-CC30 strain USA300. Beneath this is shown the oppC-trpS intergenic sequence of USA300, aligned to that MRSA 252. Asterisks above the USA300 sequence indicate stop codons of oppC and trpS. Convergent arrows indicate inverted repeats, likely comprising a rho-independent transcription terminator stem-loop structure. In all CC30 strains, the left arm of the stem is disrupted by a flanking repeat of IS1272 (shaded gray), which in turn is disrupted by ISSau2. In MRSA 252, oppB (cross-hatched) has an in-frame internal deletion that is unique to the EMRSA-16 lineage. (C) ISSau2 insertion in the genome of S. aureus TCH60 (top), which is ST30spa19 CA-MRSA (Clade 2), and corresponding segment of MRSA 252 (bottom). PCR with primers spanning the 3′-end of the 5S rRNA and flanking ilvA (right panel) reveal that this insertion is in Clade 1 strains (M809, M1015) and another Clade 2 CA-MRSA (WBG10049), but not other CC30, including strains that pre-date the Clade 1 pandemic (ATCC12598, NRS204, ATCC25923). (D) ISSau2 insertion adjacent to a 16S-23S-5S rRNA operon, and flanking okrD gene, which is unique to Clade 3 HA-MRSA. The sequence below the illustration shows the end of the 5S rRNA transcript, and adjacent rho-independent transcriptional terminator, comprised of tandem stem-loops followed by a poly-T segment. ISSau2 disrupts the right arm of the first stem-loop, with duplication of the CAT target site. (E and F) show similar disruption of a putative stem-loop structure downstream of rplQ, and a likely transcription terminator of the sbcDC operon. (G) Unique ISSau2 insertion in ST30spa33 strain MN8 disrupts stem-loop structure adjacent to saeRS regulator. The saeRS genome segment of MRSA 252 is shown for reference, and the nucleotide sequence downstream of saeS is shown above the illustration. In strain MN8, ISSau2 inserts into the left arm of a putative stem-loop structure, with duplication of the CTC target site. This is confirmed by PCR of a genomic segment spanning saeS and adjacent SAR0757, producing a 2.8 kb amplicon in MN8 (Lane 6), and a 1.2 kb product in all other strains including PT80/81 strain M1015 (Lane 1), ST30spa19 CA-MRSA strain WBG10049 (Lane 2), ST36spa16 HA-MRSA strains PM7 and PM64 (Lanes 3 and 4), and additional ST30spa33 strains UAMS-1 (Lane 5), L516 (Lane 7), and L528 (Lane 8).