Abstract
Vesicle‐mediated transport is a process carried out by virtually every cell and is required for the proper targeting and secretion of proteins. As such, there are numerous players involved to ensure that the proteins are properly localized. Overall, transport requires vesicle budding, recognition of the vesicle by the target membrane and fusion of the vesicle with the target membrane resulting in delivery of its contents. The initial interaction between the vesicle and the target membrane has been referred to as tethering. Because this is the first contact between the two membranes, tethering is critical to ensuring that specificity is achieved. It is therefore not surprising that there are numerous ‘tethering factors’ involved ranging from multisubunit complexes, coiled‐coil proteins and Rab guanosine triphosphatases. Of the multisubunit tethering complexes, one of the best studied at the molecular level is the evolutionarily conserved TRAPP complex. There are two forms of this complex: TRAPP I and TRAPP II. In yeast, these complexes function in a number of processes including endoplasmic reticulum‐to‐Golgi transport (TRAPP I) and an ill‐defined step at the trans Golgi (TRAPP II). Because the complex was first reported in 1998 (1) , there has been a decade of studies that have clarified some aspects of its function but have also raised further questions. In this review, we will discuss recent advances in our understanding of yeast and mammalian TRAPP at the structural and functional levels and its role in disease while trying to resolve some apparent discrepancies and highlighting areas for future study.
Keywords: endoplasmic reticulum, Golgi, guanine nucleotide exchange factor, sedlin, TRAPP, vesicle tethering complex
TRAPP Architecture
At present, there are 10 known yeast TRAPP subunits, and higher eukaryotes have orthologues for 8 of these (Table 1). While both yeast complexes contain the same seven subunits (Bet3p, Bet5p, Trs20p, Trs23p, Trs31p, Trs33p and Trs85p), TRAPP II is distinguished by the addition of three subunits (Trs65p, Trs120p and Trs130p). The structure of subunits common to both TRAPP I and TRAPP II has been resolved in several steps including the structures of individual subunits, followed by heterodimers. Subsequently, the structure of two mammalian subcomplexes, a heterotetramer and a heterotrimer, was resolved and docked into an electron microscopy density map of the six‐subunit TRAPP I complex. More recently, the structure of a subcomplex of five yeast subunits in complex with a guanosine triphosphatase (GTPase) was determined. Consequently, our comprehension of TRAPP architecture is the most advanced among the multisubunit complexes acting in intracellular trafficking. We will discuss insights into the structure of individual subunits and the architecture of the whole TRAPP I complex.
Table 1.
Nomenclature of yeast and mammalian TRAPP subunits
| Yeast TRAPP subunit (size in kD) | Mammalian TRAPP subunit (size in kD) | Aliases |
|---|---|---|
| Bet5p (18) | TRAPPC1 (17) | MUM‐2 |
| Trs20p (20) | TRAPPC2 (16) | Sedlin |
| Bet3p (22) | TRAPPC3 (20) | |
| Trs23p (23) | TRAPPC4 (24) | Synbindin |
| Trs31p (31) | TRAPPC5 (21) | |
| Trs33p (33) | TRAPPC6a,b (18) | |
| Trs65p (65) | None | |
| Trs85p (85) | None | |
| Trs120p (120) | TRAPPC9 (140) | NIBP |
| Trs130p (130) | TRAPPC10 (142) | TMEM‐1 |
Two protein families in TRAPP
The six different vertebrate TRAPP subunits whose structures have been solved can be divided into two families: the bet3 family composed of TRAPPC3, TRAPPC5 and TRAPPC6 and the sedlin family composed of TRAPPC1, TRAPPC2 and TRAPPC4. The low but detectable sequence similarity and the unusually high structural similarity between the three members of each family strongly suggest that they were derived from the same ancestral gene and have evolved to acquire functional diversity. Indeed, a putative TRAPPC3 ancestor has been reported in Ignicoccus hospitalis, an organism that does not possess either an endoplasmic reticulum (ER) or a Golgi (2). In yeast, the TRS33 gene is dispensable probably because of the formation of a homodimer of Bet3p that can partly replace Bet3p–Trs33p in TRAPP I (3). Structural similarity between subunits has also been recently reported for another tethering complex called exocyst. In that case, structural similarity was seen between long α‐helical rod‐like structures of Exo70p and the C‐terminal domains of Sec6p, Exo84p and Sec15 4, 5. Furthermore, structural similarities between subunits of the conserved oligomeric Golgi (COG) vesicle tethering complex have been speculated as well 4, 6, 7. Thus, structural similarities between the subunits of tethering factors appear to be an emerging common theme. Like the TRAPP subunits, each subunit in the other tethering complexes would play a distinctive role.
Structural features of TRAPP subunits
The structure of TRAPPC1–TRAPPC3–TRAPPC4–TRAPPC6a revealed that TRAPPC4 contains an internal insertion of a domain missing from its yeast orthologue Trs23p. This domain exhibits a barely detectable sequence homology with several PDZ domains, the highest similarity being with the sixth PDZ domain of InaD‐like protein. However, the sequence similarity is limited to the C‐terminal 40 residues, as observed by others (8). Furthermore, in TRAPPC4, this domain does not contain the Gly‐Leu‐Gly‐Phe signature sequence of classical PDZ domains. It is therefore referred to as the PDZ‐like (PDZL) domain. The PDZL domain is present in Caenorhabditis elegans and higher organisms (9). Because the PDZ domain is a protein‐interacting module and because the PDZL domain in the tetrameric complex contains a characteristic surface groove, it is likely to bind an internal sequence of an as‐yet‐unknown protein. The lack of interaction between the two vertebrate recombinant TRAPP I subcomplexes (see below) and the presence of the PDZL domain in TRAPPC4 are the two most unique features of the vertebrate complex when compared with yeast TRAPP I.
TRAPPC2 is structurally quite similar to the N‐terminal regulatory domain of two SNARE proteins Ykt6p and Sec22b (10). The TRAPPC2 structure has a surface‐exposed residue Asp47 on the α1 helix, the mutation of which to tyrosine causes the genetic disease spondyloepiphyseal dysplasia tarda (SEDT) (11). The corresponding region in the N‐terminal regulatory domain of Ykt6p was implicated in the binding of its C‐terminal SNARE domain (12) to form a so‐called closed conformation that inhibits intermolecular SNARE complex formation 12, 13. This surface is composed of highly conserved residues that are exposed in the context of the TRAPPC2–TRAPPC3–TRAPPC5 subcomplex (9). One possibility is that this region binds a SNARE domain to regulate SNARE complex formation, thus linking TRAPP to SNAREs as has been reported for other vesicle tethering complexes 14, 15, 16, 17.
The crystal structure of TRAPPC3 and, more recently of Bet3p, revealed an interesting structural feature in which a hydrophobic channel is formed within the core of the protein that is accessible from the surface 18, 19. This hydrophobic channel is quite unique, and such a channel has only been reported in one other protein, ORF‐9b, of the SARS coronavirus (20). The function of this feature is unknown, but a mutation in the yeast protein (bet3‐4), which is composed of an A94L mutation within the channel and a carboxy terminal hemagglutinin tag, is temperature sensitive for growth and localizes to multiple membranes within the cell (19). Cai et al. confirmed the growth phenotype and further demonstrated that it is the combination of these two changes to the protein that accounts for the growth defect, suggesting an influence of the carboxy terminus on the function of the channel. This mutant underscores the fact that this channel plays some uncharacterized but critical role in the function of TRAPPC3/Bet3p. Heinemann and co‐workers (21) suggest that palmitoylation of TRAPPC3 allows for the insertion of an acyl chain into the channel, which stabilizes the structure of the protein. However, TRAPPC5/Trs31p and TRAPPC6/Trs33p, the two other TRAPP subunits that are structural homologues of TRAPPC3 9, 18, 19, 22, do not have such a channel, and it is unclear why only the TRAPPC3 protein would have it. Furthermore, mutation of the cysteine residue that is acylated in the recombinant TRAPPC3/Bet3p protein to a serine blocks acylation and displays no growth phenotype in yeast 19, 23. This strongly argues that there must be another purpose to this channel. One possibility suggested by Kim et al. (19) is that an acyl chain on an unidentified Golgi protein is inserted into the channel. Perhaps, the carboxy terminus of TRAPPC3/Bet3p is somehow involved in this insertion, providing an explanation as to why both modifications to the Bet3p protein are required to generate the phenotype of the bet3‐4 allele.
A detailed view of TRAPP I
The architecture of the TRAPP I complex was assembled by first resolving the structures of the mammalian tetrameric TRAPPC1–TRAPPC3–TRAPPC4–TRAPPC6a and the trimeric TRAPPC2–TRAPPC3–TRAPPC5 subcomplexes (9). In these two subcomplexes, TRAPPC3 interacts with TRAPPC6 or TRAPPC5 using a common surface, and each heterodimer interacts with another subunit side by side to form flat subcomplexes. While the two mammalian subcomplexes do not interact with each other, the corresponding subcomplexes of yeast TRAPP I form a single tight complex when the six subunits were coexpressed (9). Single particle electron microscopy of this complex revealed that yeast TRAPP I has an elongated, flat architecture. Into the ∼30 Å resolution three‐dimensional reconstituted image, the two structures of the mammalian subcomplexes could be nicely docked, providing a pseudo high‐resolution structure of TRAPP I (Figure 1B) (9). More recently, a heteropentameric yeast TRAPP I assembly (Bet3p–Trs31p–Trs23p–Bet5p–Bet3p) in complex with Ypt1p was reported (Figure 1A) (18).
Figure 1.
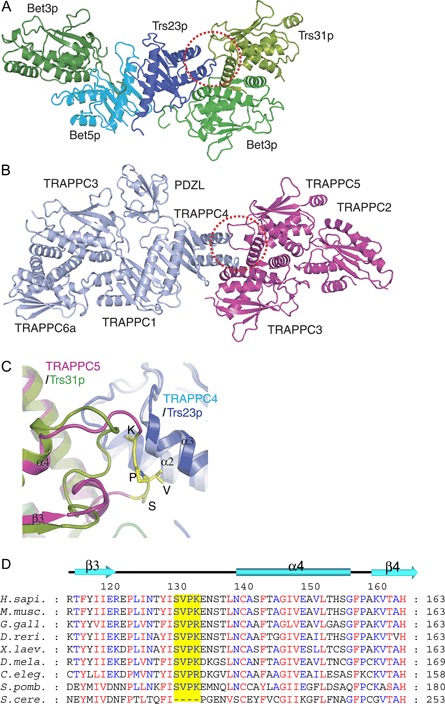
The structure of yeast and mammalian TRAPP subcomplexes. A) The structure of a subcomplex of yeast TRAPP containing two copies of Bet3p and one each of Bet5p, Trs23p and Trs31p in complex with the GTPase Ypt1p was solved to 3.7 Å resolution (18). The TRAPP subunit models are shown in the absence of Ypt1p that would be on the surface facing the reader. B) The structure of two subcomplexes of mammalian TRAPP were solved to 2.4 Å resolution (TRAPPC1–TRAPPC3–TRAPPC4–TRAPPC6a, slate color) and 2.1 Å resolution (TRAPPC2–TRAPPC3–TRAPPC5, pink color) (9). The main differences between the yeast and the mammalian complexes are the presence of a PDZL domain on TRAPPC4 and an insertion in the β3‐α4 loop of TRAPPC5, resulting in a protrusion at the TRAPPC4–TRAPPC5 interface (circled in panels A and B). C) The TRAPPC5–TRAPPC4 interface is superimposed onto the Trs31p–Trs23p interface with the protruding region of TRAPPC5 colored in yellow. Panels (A–C) were rendered in PyMol. D) A multi‐sequence alignment of TRAPPC5/Trs31p orthologues. The red and blue letters indicate the amino acids that are 100 and >70% conserved in nine representative orthologues. Secondary structural elements are indicated above the alignment. The ‘SVPK’ insertion in the β3‐α4 loop of TRAPPC5 is highlighted in yellow. Aligned sequences are H. sapi. (Homo sapiens), M. musc. (Mus musculus), G. gall. (Gallus gallus), D. reri. (Dario rerio), X. laev. (Xenopus laevis), D. mela. (Drosophila melanogaster), C. eleg. (Caenorhabditis elegans), S. pomb. (Schizosaccharomyces pombe) and S. cere. (Saccharomyces cerevisiae).
In the context of the TRAPP complex, TRAPPC2 is situated at one end, while the TRAPPC3–TRAPPC6 heterodimer is on the opposite end (Figure 1B). The TRAPPC1 and TRAPPC4 subunits bridge the two ends of the complex and are required for interaction with the GTPase Ypt1p (see below). The positioning of TRAPPC2 is interesting as it is not involved in nucleotide exchange activity (9) or vesicle tethering (24) (see below). Therefore, the exposed relatively large surface area is available for potential interactions with other proteins involved in membrane transport as discussed above. The arrangement of the subunits within the complex is such that two large (∼180 × 65 Å) flat surfaces are presented. Part of one or both of these surfaces will be occupied by the TRAPP II‐specific subunits, but exactly where they interact with this core of the TRAPP complex is still unknown. A recent study suggests that they overlap with the Ypt1p‐binding site (25). Now that the site of interaction between TRAPP I and Ypt1p has begun to be elucidated (18) (Figure 3); this theory can be put to the test.
Figure 3.
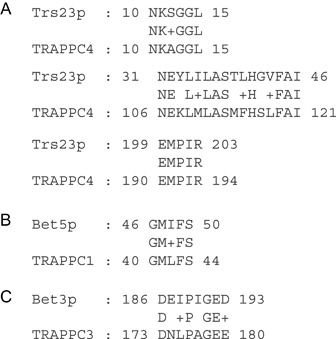
Conservation of residues in TRAPP I required for Ypt1p contact. The amino acid sequences at the contact patches between Ypt1p and TRAPP I are highly conserved between yeast and humans. Shown are sequence alignments for the three relevant TRAPP I subunits using BLAST of A) yeast Trs23p with human TRAPPC4, B) yeast Bet5p with human TRAPPC1 and C) yeast Bet3p with human TRAPPC3.
TRAPP as a guanine nucleotide exchange factor
Ypt/Rab GTPases are conserved key controllers of the different protein transport steps in all eukaryotic cells. They switch between the GTP‐bound ‘on’ and the GDP‐bound ‘off’ states with the help of upstream regulators. Guanine nucleotide exchange factors (GEFs) activate GTPases by accelerating their intrinsic GDP release and GTP uptake reactions. While in the on state, Ypt/Rabs interact with multiple effectors that mediate the various substeps of vesicular transport, from vesicle formation through their motility, tethering and fusion. A number of Ypt/Rab GEFs have been identified (26), and some of these are multiprotein complexes. While examples exist for GEF complexes that also contain an effector for the same Ypt/Rab 27, 28, TRAPP is the only complex that has been implicated in activation of more than one Ypt/Rab: Ypt1p and Ypt31/32p.
TRAPP I as a GEF for Ypt1p
The Ypt1p GTPase regulates the ER‐to‐Golgi transport step 29, 30, 31. Independent characterization of the Ypt1‐GEF and the TRAPP complex from yeast cells revealed a surprising similarity between the biochemical properties as well as the cellular localization of the two complexes 1, 32, 33. Indeed, the two complexes turned out to be identical, and TRAPP I was shown to act as a Ypt1p GEF by the two groups 34, 35. TRAPP I purified from yeast cells accelerates both GDP release and GTP uptake by Ypt1 34, 36 with reported values for recombinant TRAPP of >400‐ and 30‐fold, respectively (18). These values are similar to those reported for other known GEFs.
Findings regarding which TRAPP I subunit(s) act as the Ypt1p GEF turned out to be surprising as well. Three of the known Rab GEFs consist of a single protein: the Vam6p/Vps39p subunit of the homotypic fusion and vacuole protein sorting (HOPS) complex acts as a GEF for Ypt7p (37), Sec2p acts as a GEF for Sec4p (38) and Vps9p acts as a GEF for Vps21/Ypt51p (39). The Ypt6p‐GEF requires two proteins for its activity, Ric1p and Rgp1p (40). In contrast, Kim et al. (9) showed that the minimal Ypt1p‐GEF complex consists of four TRAPP subunits essential for cell viability: Bet3p, Bet5p, Trs23p and Trs31p.
The structure of Ypt1p in complex with the minimal TRAPP I GEF at 3.7 Å resolution was recently solved (18). In this structure, Ypt1p is proximal to three domains of Trs23p (residues 10–15, 31–46 and 199–203), one α‐helix of Bet5p and the C‐termini of both of the Bet3p subunits present in this complex (referred to as Bet3p‐A and Bet3p‐B) (Figure 2). Based on the difference between the five‐subunit structure and the previously published individual subunit structures, the authors proposed that Trs31p does not interact with Ypt1p but is important for the formation of the Ypt1p–TRAPP I interface. The side view of the published structure (Figure 2A) together with analysis of putative TRAPP I–Ypt1p interactions using a 4.0 Å distance cut‐off fails to unequivocally identify the critical residues that stabilize the complex. However, sequence analysis of the proposed Ypt1p contact patches on Trs23p, Bet5p and Bet3p shows that all the patches are remarkably conserved from yeast to humans (Figure 3). While the patch conservation supports the structural model, finer structure resolution and binding analysis of Ypt1p with relevant TRAPP I subunit mutations are required for nailing down the specific interactions.
Figure 2.
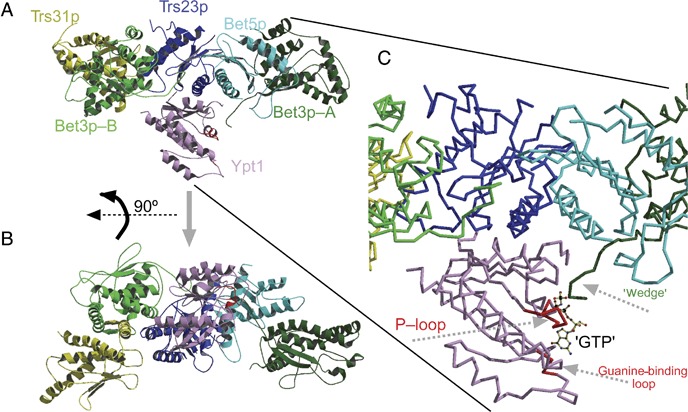
The TRAPP I–Ypt1p interface. Ypt1p–TRAPP I contact regions span three TRAPP I subunits as revealed by the Ypt1p–TRAPP I complex recently reported by Cai et al. (18). A) Ribbon diagram of the Ypt1p–TRAPP I complex. B) A 90° rotated view relative to that shown in A). The orientation is similar to that shown in Cai et al. C) A close‐up view showing the important putative interactions between TRAPP I and Ypt1. To demonstrate the incompatibility between the C‐terminal region (wedge) of Bet3p‐A with the presence of nucleotide on Ypt1p when in complex with TRAPP, a GTP analog (GppNHp) was docked to the Ypt1p–TRAPP I complex based on its position in the structure of Ypt1p‐GTP (PDB ID 1YZN). Note how residues from the C‐terminus of Bet3p‐A occupy a similar position to that of the phosphate groups of the nucleotide. Figure prepared with Molscript.
The general model for the mechanism by which GEFs stimulate GDP release involves conformational changes in the two switch domains and the P‐loop of the GTPase (41). Based on the Ypt1p–TRAPP I structure, Cai et al. (18) propose a mechanism for stimulation of GDP release from Ypt1p by TRAPP I. First, they propose that three of the five TRAPP subunits in the complex are important for Ypt1p binding: namely Trs23p, Bet5p and Bet3p. Second, they suggest that Bet5p links Trs23p and Bet3p‐A and is important for directing the C‐terminus of Bet3p‐A to the Ypt1p nucleotide‐binding pocket. Analysis of mutations in these domains in Trs23p and Bet5p supports their importance for GEF activity 9, 18. However, it is not clear yet whether these mutations affect complex assembly, Ypt1p binding or GDP release. Finally, Cai et al. assign the critical catalytic activity to the C‐terminus of Bet3p‐A, proposing that it is inserted into the GTP‐binding pocket of Ypt1p and functions as a ‘wedge’ to initiate the conformational change required for GDP release. Indeed, when a GTP molecule is docked into the published structure (Figure 2C), it is clear that residues from the C‐terminus of Bet3p‐A occupy a similar position to that of the phosphate groups of the nucleotide. However, mutations altering the charge have no effect on GEF activity, and deletion of the whole C‐terminus does not affect cell growth. In addition, the tested Bet3p mutations might instead affect the second Bet3p molecule, Bet3p‐B, and its interaction with Trs23p and Ypt1p. Thus, the mechanistic aspect of the GEF activity of TRAPP will have to be addressed in future studies.
TRAPP II as a GEF for Ypt31/32p
The Ypt31/32p GTPase functional pair acts at the trans Golgi (42) as does the TRAPP II complex 36, 43, 44. TRAPP purified from yeast lysates was shown to act as a GEF for Ypt31/32p (34). This TRAPP contains a mixture of TRAPP I and TRAPP II, and further purification revealed the TRAPP II complex exhibiting this activity. Moreover, mutations in the two essential TRAPP II‐specific subunits, Trs120p and Trs130p, abolish the Ypt31/32p GEF activity of TRAPP while increasing the Ypt1p GEF activity (25). This last result shows that the TRAPP II‐specific subunits are required for the Ypt31/32p GEF activity and for the inhibition of the Ypt1p GEF activity of TRAPP II. This biochemical activity has been controversial in the field because of discrepant results between two research groups 18, 35, 45. This may be because of differences in complex purification and GEF assay conditions. Because TRAPP purified from yeast lysates contains both complexes, it is possible that conditions used by the Ferro‐Novick group during purification or assaying of the GEF activity favor dissociation of the TRAPP II‐specific subunits, thus yielding mostly TRAPP I that has only a Ypt1p‐GEF activity. Alternatively, it is possible that TRAPP II purified by the Segev group contains additional unknown subunits required for the Ypt31/32p GEF activity. This controversy will have to await reconstitution of the Ypt31/32p GEF activity using recombinant TRAPP II subunits.
Physical, genetic and cellular studies support a role for TRAPP I and TRAPP II as GEFs for Ypt1p and Ypt31/32p, respectively. First, the TRAPP II‐specific subunit Trs130p interacts with the nucleotide‐free form of Ypt31p, but not of Ypt1p, in a yeast two‐hybrid assay. Second, the BET3 gene encoding the TRAPP I/II subunit Bet3p interacts synergistically with YPT1 and YPT31/32 (25), whereas the genes encoding TRAPP II‐specific subunits, TRS120 and TRS130, were shown to interact with YPT31/32, but not with YPT1, in four independent studies 45, 46, 47, 48. Finally, Trs130p regulates the intracellular distribution of Ypt1p and Ypt31/32p in opposite ways. Specifically, in trs130 mutant cells, Ypt31/32p staining is diffuse, while Ypt1p staining is enhanced (25). Together, these observations support the biochemical results suggesting that TRAPP II is a Ypt31/32p‐GEF. Based on the conclusion that TRAPP I preferentially acts as a Ypt1p GEF and TRAPP II acts as a Ypt31/32p GEF, Morozova et al. (25) proposed that sequential activation of the Ypts that control Golgi entry and exit by TRAPP I and TRAPP II co‐ordinates these two transport steps. The role of the TRAPP complexes in co‐ordination of transport steps is an important future question in the field.
Is mammalian TRAPP a Rab‐GEF?
A Rab‐GEF activity of the mammalian TRAPP has not been shown yet. The two bacterially expressed reconstituted vertebrate subcomplexes do not exhibit GEF activity either alone or in combination (unpublished data). We speculate that the reason for this lack of activity is because the GEF activity of the reconstituted yeast TRAPP I requires the presence of four subunits, Bet3p, Trs31p, Bet5p and Trs23p, in one complex, whereas their vertebrate counterparts are present on the two separate subcomplexes. Therefore, one obvious question to be addressed is why would the vertebrate subcomplexes not readily associate with each other? In the yeast TRAPP I holocomplex, Bet3p–Trs31p forms an interface for binding Trs23p, thereby linking the Bet3p–Trs20p–Trs31p and Bet3p–Trs33p–Bet5p–Trs23p subcomplexes together. At the three‐way interface, the β3‐α4 loop of Trs31p has a conformation largely different from that of TRAPPC5 in the TRAPPC2–TRAPPC3–TRAPPC5 subcomplex (Figure 1C) (18). A sequence alignment reveals that vertebrate TRAPPC5 contains a unique insertion of the amino acid residues SVPK compared with yeast Trs31p (Figure 1D). We suggest that this insertion may be responsible for preventing the two vertebrate subcomplexes from interacting with each other. It is noteworthy that the SVPK sequence is also found in Schizosaccharomyces pombe Trs31p, however the assembly state of TRAPP in this organism is presently unknown. It is possible that an additional unknown subunit bridges the two vertebrate subcomplexes to form a catalytically active complex. It is also possible that a posttranslational modification is required for the assembly and/or the Rab‐GEF activity of the vertebrate TRAPP. Alternatively, vertebrate TRAPP I may not have GEF activity at all, and a yet unknown protein is present in vertebrate cells to activate the Ypt1p orthologue Rab1. So far, TRAPP I is the only known GEF for Ypt1p. Based on the conservation of Ypt/Rabs and TRAPP in general and specifically the remarkable conservation of the proposed Ypt1p contact patches on the TRAPP I subunits (Figure 3), we expect that the mammalian TRAPP complex acts as a Rab1‐GEF.
A role for TRAPP in vesicle tethering
In addition to the well‐established role for TRAPP in Ypt activation, both TRAPP I and TRAPP II were implicated in vesicle tethering.
Yeast TRAPP as a tether
TRAPP I plays an essential role in ER‐to‐Golgi transport 1, 36, 49, 50. Its role as a tether in this transport step was established by showing that TRAPP I interacts directly with ER‐derived vesicles produced using either a crude cytosolic fraction or purified COP II coat components (36). How does TRAPP I interact with vesicles? It was recently reported that ER‐derived vesicles bind to TRAPP I through an interaction between the Bet3p subunit and the vesicle coat protein Sec23p (24). This was a surprising result for two reasons. First, it was long assumed that vesicles rapidly uncoat following inactivation of Sar1p by hydrolysis of bound GTP (51). This uncoating is catalyzed by the GTPase‐activating protein activity of Sec23p towards Sar1p (52). Using a liposome system, Sato and Nakano (53) showed that the Sec23p/Sec24p complex remains associated with prebudding complexes because of constant reactivation of Sar1p by its GEF, Sec12p. Although Sec12p is not found in transport vesicles in Saccharomyces cerevisiae, other factors may stabilize the association of Sec23p with free vesicles as has been suggested to happen in Pichia pastoris and higher eukaryotes 54, 55. Second, an earlier study showed that TRAPP I can bind to COP II vesicles in vitro, but the interaction was blocked by the inclusion of a nonhydrolyzable GTP analog (36). The latter result was interpreted as a requirement for vesicle uncoating before the TRAPP I interaction. If, however, sufficient amounts of Sec23p remain associated with free vesicles, then the previous result may reflect the fact that other GTPases are involved in the tethering of these vesicles. In agreement with this idea, it has been shown that the protein GDI interferes with vesicle tethering (56) and GDI binds to the non‐activated (GDP‐bound) form of the GTPase Ypt1p. However, Ypt1p activation is necessary for vesicle fusion (57). Because Ypt1p is reportedly associated with vesicles (58), it is plausible that premature activation of Ypt1p interferes with vesicle tethering and its GDP‐bound form participates in tethering. In this scenario, activated Ypt1p would recruit other factors to the site of the tethered vesicle to further strengthen the tether or aid in SNARE complex assembly.
Still unresolved is why ER‐derived vesicles would not interact with the related TRAPP II complex that also contains two copies of Bet3p. One possibility is that the TRAPP II‐specific subunits block the site of interaction between Bet3p and Sec23p. A recent study suggests that the TRAPP II subunit Trs120p is required for the stability of the Trs130p subunit, suggesting that these two proteins might interact (25). Therefore, it seems unlikely that Trs120p and Trs130p independently block both copies of Bet3p from interacting with Sec23p. Furthermore, Uso1p has been reported to be involved in the initial tethering of vesicles to the Golgi in yeast (56) and proteins in mammalian cells such as p115 and GM130 are also involved in this process 59, 60. A likely possibility is that there are multiple means for a vesicle to be tethered to the Golgi involving TRAPP, Uso1p, coat proteins and most likely other proteins, each being critical to the process such that disruption of one of these interactions weakens the overall vesicle–membrane interaction.
While the bulk of studies on yeast have focused on the smaller TRAPP I complex, given the relatedness between TRAPP I and TRAPP II, it will be important to more fully characterize the larger complex. Several studies have begun to address the transport step in which TRAPP II functions. Mutants in yeast, trs130 and trs120, were shown to accumulate early Golgi forms of invertase. In addition, the trs130 mutants also accumulated early Golgi forms of carboxypeptidase Y as well as Berkeley bodies, all suggestive of a block in traffic beyond the ER‐to‐Golgi step (36). Interestingly, a subsequent study in which a variety of truncations in trs120 were examined showed that some alleles affected traffic between the endosome and the trans Golgi (43). Not all proteins that rely on a functional endosome‐to‐trans Golgi pathway were affected in these mutants. For example, carboxypeptidase Y was not affected in trs120‐2, trs120‐4 and trs120‐8, yet other proteins such as Snc1p–green fluorescent protein (GFP) were blocked. This led to the conclusion that Trs120p functions in traffic between the early endosome and the trans Golgi, a notion that fits well with its GEF activity for Ypt31/32p (61). However, the same study showed that mutations in trs130 can block secretion in general and affect localization of both carboxypeptidase Y and Snc1p–GFP (43). This would indicate a complex function for TRAPP II in multiple pathways, and clarification of its function awaits further studies. In addition, it has been shown that the Golgi‐associated retrograde protein (GARP) tethering complex also functions at the trans Golgi (62), and understanding the differing roles of TRAPP II and GARP will be of interest. The TRAPP II‐specific Trs65p subunit has no known mammalian orthologue (Table 1) and is presumed to have a fungal‐specific function. While it has been implicated in cell wall biogenesis and stress response, its role in TRAPP II is supported by the findings that the protein colocalizes with Trs130p and deletion of TRS65 in yeast leads to a conditional lethal phenotype if either one of the other TRAPP II‐specific subunits is modified (44). Furthermore, the trs65 mutant has reduced Ypt31/32p GEF activity.
Whereas TRAPP I was implicated in binding the COP II coat subunit Sec23p (see above), TRAPP II was reported to interact with the COP I vesicle coat (43). Together, these findings suggest that the two TRAPP complexes can distinguish between various vesicle coat proteins. Yet, COP I vesicles have been shown to interact with several other vesicle tethering complexes including COG (63) and Dsl1 (64), and, as stated above, the Bet3p subunit that interacts with Sec23p is also present in TRAPP II. Clearly, there must be other components of the vesicles that interact with each of these complexes to allow for a distinction between compartments to be made. It is possible that an initial interaction between these complexes and coat proteins occurs followed by a second even more specific interaction.
Mammalian TRAPP as a tether
The study of mammalian TRAPP lags behind that of the yeast complexes but has begun to be addressed. The subunits are widely expressed across many different tissues consistent with a ubiquitous function for this complex 8, 65, 66, 67, 68, 69. Unlike its yeast counterpart, the subunits of the mammalian complex are largely found in an unassembled state 68, 70, a finding that is supported by an immunofluorescence study on the TRAPPC3 subunit (68). Interestingly, a subsequent study on TRAPPC3 showed clear localization at transitional ER sites (71), perhaps reflecting subtle differences in sample preparation. An in vitro assay reconstituting ER‐to‐Golgi transport in NRK cells showed that the TRAPPC3 subunit functions before both the GTPase Rab1 and the SNARE‐binding protein α‐SNAP but after the COP II vesicle budding step (68), similar to the yeast TRAPP I complex.
One of the main differences between yeast and mammalian complexes is in their reported functions. The mammalian complex was proposed to be involved in homotypic fusion of ER‐derived COP II vesicles (71). This was based on the localization of the TRAPPC3 subunit to transitional ER sites and the inhibition of COP II vesicle fusion in an in vitro assay that reconstitutes this event. Given the well‐documented interaction between COP II vesicles and Golgi in yeast, this would imply that the mammalian TRAPP complex functions by a different mechanism than its yeast counterpart. It is noteworthy, however, that the same antibody used to interfere with the homotypic fusion assay was reported to inefficiently precipitate the native form of the protein (24). This raises the question as to how such an antibody can interfere in an assay that presumably requires native TRAPPC3. One possibility is that a TRAPPC3 epitope not normally exposed in the context of the entire complex is exposed in the homotypic fusion assay. This would imply that only TRAPPC3, and not the TRAPP complex per se, is involved in this tethering event. Future studies should be aimed at resolving this issue.
Molecular models for TRAPP as a tether
Thus far, two models for the orientation of the TRAPP complex on membranes have been proposed. In one (9), the complex is proposed to lay flat on Golgi membranes and interact with an incoming vesicle. A second model (18) suggests that the complex does not lay flat upon membranes but rather the two copies of the TRAPPC3/Bet3p subunit bridge two membranes. The main difference between the models is that the first model seeks to explain heterotypic fusion, while the latter model seeks to explain homotypic fusion, a function that may be specific to the mammalian complex or to TRAPPC3. Indeed, Cai et al. (18) speculate that the arrangement of the complex with respect to membranes is similar between the two types of fusion events, a notion that is as yet unproven. It is entirely possible that the arrangement of the complex with respect to membranes is as different as the fusion events themselves (i.e. homotypic versus heterotypic). Evidence suggesting possible differing functions for the subunits in yeast and mammals is the fact that while two forms of TRAPP have been described in yeast, only a single high‐molecular‐weight peak on a gel filtration column has been reported in mammalian cells 68, 70. Furthermore, homotypic fusion has not been reported in yeast. In that case, these models are not necessarily mutually exclusive and indeed may reflect the evolutionary needs of the complex to mediate two different types of fusion events. As more work on TRAPP is performed, it is likely that refinement of both models will be needed.
Other possible functions
TRAPP subunits have also been linked to other cellular processes that are briefly reviewed in this study. While some of the data relate to membrane transport steps other than those described above, there are also data to suggest functions in different cellular processes.
Along with a well‐established role for TRAPPC2 in SEDT (see below), this subunit has also been reported to regulate gene transcription 66, 72 and to weakly interact with chloride intracellular ion channels 1 and 2 73, 74. While the latter may be related to a role in trafficking, the former is presumably not.
TRAPPC4 was identified as a protein called synbindin and was shown to interact with the cell surface protein syndecan‐2 (8). This interaction, reported to be mediated by the PDZL domain of TRAPPC4, is involved in the formation of structures called dendritic spines. As the strength of the interaction between TRAPPC4 and syndecan‐2 is not known, it remains unclear as to whether the observed morphological defect is the result of a direct interaction between the proteins or some sort of indirect effect of early secretory traffic on dendritic spines.
A mutation in TRAPPC6a leads to a mosaic loss of coat pigment likely because of a defect in melanosome formation (75). This might indicate either that TRAPPC6a has a specific melanocyte function or that a defect in early secretory protein traffic interferes with melanosome formation (76). In the latter possibility, it is unclear why such a specific defect would be seen.
The Trs85p subunit was first identified as functioning in sporulation (77). It was subsequently shown to be a component of TRAPP and to function in ER‐to‐Golgi transport (36). Recently, Trs85p was reported to function in the cytosol‐to‐vacuole targeting pathway, suggesting a role for this subunit in autophagy as well as in secretion 78, 79.
TRAPP II‐specific subunits have also been implicated in other processes. NIBP is a mammalian protein displaying the highest degree of homology with yeast Trs120p (80), although it was not detected by precipitation of TRAPPC3 (81). The protein is well conserved through evolution 80, 82 and interacts with NIK and IKKβ, which activates the transcription factor NF‐κB (67). Curiously, knockdown of NIBP prevented nerve growth factor‐induced neurite extension in PC12 cells. Whether this latter effect is because of a direct role of NIBP in NF‐κB signaling in neuronal cells or because of the fact that neurite outgrowth requires additional membranes and, therefore, a more active secretory pathway will need to be addressed.
In yeast, a trs130 mutation leads to elevated expression of OCH1, a gene whose product is required for initiation of mannose outer chain elongation (46). Together with the links between transcription and both TRAPPC2 and NIBP (see above), this would be the third TRAPP subunit tied to this process.
Clearly, all these studies collectively suggest that either there is a yet‐to‐be explored link between TRAPP biology and other cellular processes or TRAPP subunits can fulfill multiple, unrelated roles.
TRAPP and disease
At least two TRAPP I subunits have been implicated in human diseases: TRAPPC1 and TRAPPC2. Mutations in TRAPPC1 (MUM‐2) were reported to result in expression of antigenic peptides in a melanoma and may cause increased immune response associated with this disease (83), but this remains a single case report.
Mutations in TRAPPC2 (sedlin) were linked to SEDT (65) and have been more thoroughly characterized. This disorder primarily affects the epiphyses and manifests in early adolescent males. Affected individuals suffer from back pain and dysplasia of the large joints and are below the average family height, the latter being because of a flattening of the vertebrae. Subsequent studies have linked numerous mutations in TRAPPC2 to SEDT including missense mutations, truncations and RNA splicing mutations (84). It has been reported that patients with this disorder have extracellular collagen fibrils that are shorter and more frayed than unaffected patients (85), leading to the hypothesis that a defect in collagen trafficking in chondrocytes is responsible for the disorder 85, 86. However, several pieces of evidence suggest that the role of TRAPPC2 in SEDT may not be that simple. First, there is no evidence that patients suffering from SEDT have any other membrane traffic defects. This is unexpected considering that TRAPPC2 is widely expressed. Second, if TRAPPC2 is only involved in collagen trafficking, one would expect defects in other collagen‐secreting cells. Again, this has also not been reported. Finally, one study showed that mutated TRAPPC2 accumulated in the nucleus (87), suggesting that TRAPPC2 may have an undescribed nuclear function that should be taken into account when seeking to describe the tissue‐specific defect of SEDT. In this respect, it is noteworthy that TRAPPC2 has been identified as MIP‐2A, a protein involved in the regulation of gene transcription 66, 72. While it has been suggested that TRAPPC2 is involved in guanine nucleotide exchange based on its structural similarity to signal recognition particle receptor β(88), it has recently been demonstrated that its yeast orthologue, Trs20p, is not necessary for the Ypt1p‐directed nucleotide exchange function (9). How then is TRAPPC2 involved in the etiology of SEDT? This remains an open question, but a particular mutation could eventually be revealing. One affected patient has an Asp47→Tyr mutation that, based on the structure of TRAPPC2 in the context of the TRAPP complex, is exposed on the surface of the protein and is not involved in any interaction between TRAPP subunits (9). This particular mutation does not efficiently compensate for the loss of Trs20p in yeast, whereas wild‐type TRAPPC2 does (89) suggesting a conserved function for this residue. Indeed, Asp47 is highly conserved 10, 89. Therefore, identification of TRAPPC2‐interacting partners, particularly those that are affected by mutation of Asp47, will be an important step in understanding the link between TRAPPC2 and SEDT.
Concluding remarks
Progress has been made in understanding how TRAPP fits into the overall process of Ypt/Rab activation and vesicle tethering at the different ends of the Golgi. Yet these studies have led to numerous new questions. Particularly interesting will be to understand how the yeast work relates to the more complex higher eukaryotic early secretory pathway. Are the lessons learned in one model system readily translatable to the other? Why are two complexes readily identified in yeast but not in mammalian cells? How is TRAPP’s role in heterotypic fusion to be reconciled with its reported role in homotypic fusion? Added complexities of higher eukaryotes include an intermediate compartment between the ER and Golgi (referred to as ERGIC) and TRAPP subunit isoforms for at least the TRAPPC6 subunit. This may all point to related but more specialized functions for this complex in mammals. Also of interest, mechanistically will be to address how the Ypt‐GEF activity of TRAPP is related to its role as a tether. With 10 years of research, TRAPP biology has emerged from its infancy and has turned into an exciting area of study. As TRAPP enters its ‘teens’ and proceeds through its second decade of research, there is every reason to believe the findings will be just as interesting.
Acknowledgments
Work in the authors’ laboratories is supported by Canadian Institutes of Health Research, Natural Sciences and Engineering Research Council of Canada and the Canada Foundation for Innovation (M. S.); 21C Frontier Microbial Genomics and Applications Center Program, Ministry of Education, Science & Technology, Republic of Korea (Y‐G. K.); Creative Research Initiatives (Center for Biomolecular Recognition) of MOST/KOSEF (B‐H. O.); National Institutes of Health (N. S.). We are grateful to members of our laboratories for helpful comments and discussions on this manuscript.
References
- 1. Sacher M, Jiang Y, Barrowman J, Scarpa A, Burston J, Zhang L, Schieltz D, Yates JR III, Abeliovich H, Ferro‐Novick S. TRAPP, a highly conserved novel complex on the cis‐Golgi that mediates vesicle docking and fusion. EMBO J 1998;17:2494–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Podar M, Wall MA, Makarova KS, Koonin EV. The prokaryotic V4R domain is the likely ancestor of a key component of the eukaryotic vesicle transport system. Biol Direct 2008;3:2–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim MS, Yi MJ, Lee KH, Wagner J, Munger C, Kim YG, Whiteway M, Cygler M, Oh BH, Sacher M. Biochemical and crystallographic studies reveal a specific interaction between TRAPP subunits Trs33p and Bet3p. Traffic 2005;6:1183–1195. [DOI] [PubMed] [Google Scholar]
- 4. Dong G, Hutagalung AH, Fu C, Novick P, Reinisch KM. The structures of exocyst subunit Exo70p and the Exo84p C‐terminal domains reveal a common motif. Nat Struct Mol Biol 2005;12:1094–1100. [DOI] [PubMed] [Google Scholar]
- 5. Sivaram MV, Furgason ML, Brewer DN, Munson M. The structure of the exocyst subunit Sec6p defines a conserved architecture with diverse roles. Nat Struct Mol Biol 2006;13:555–556. [DOI] [PubMed] [Google Scholar]
- 6. Cavanaugh LF, Chen X, Richardson BC, Ungar D, Pelczer I, Rizo J, Hughson FM. Structural analysis of conserved oligomeric Golgi complex subunit 2. J Biol Chem 2007;282:23418–23426. [DOI] [PubMed] [Google Scholar]
- 7. Whyte JR, Munro S. Vesicle tethering complexes in membrane traffic. J Cell Sci 2002;115:2627–2637. [DOI] [PubMed] [Google Scholar]
- 8. Ethell IM, Hagihara K, Miura Y, Irie F, Yamaguchi Y. Synbindin, a novel syndecan‐2‐binding protein in neuronal dendritic spines. J Cell Biol 2000;151:53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim YG, Raunser S, Munger C, Wagner J, Song YL, Cygler M, Walz T, Oh BH, Sacher M. The architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell 2006;127:817–830. [DOI] [PubMed] [Google Scholar]
- 10. Jang SB, Kim YG, Cho YS, Suh PG, Kim KH, Oh BH. Crystal structure of SEDL and its implications for a genetic disease spondyloepiphyseal dysplasia tarda. J Biol Chem 2002;277:49863–49869. [DOI] [PubMed] [Google Scholar]
- 11. Gedeon AK, Tiller GE, Le Merrer M, Heuertz S, Tranebjaerg L, Chitayat D, Robertson S, Glass IA, Savarirayan R, Cole WG, Rimoin DL, Kousseff BG, Ohashi H, Zabel B, Munnich A et al The molecular basis of X‐linked spondyloepiphyseal dysplasia tarda. Am J Hum Genet 2001;68:1386–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tochio H, Tsui MM, Banfield DK, Zhang M. An autoinhibitory mechanism for nonsyntaxin SNARE proteins revealed by the structure of Ykt6p. Science 2001;293:698–702. [DOI] [PubMed] [Google Scholar]
- 13. Nicholson KL, Munson M, Miller RB, Filip TJ, Fairman R, Hughson FM. Regulation of SNARE complex assembly by an N‐terminal domain of the t‐SNARE Sso1p. Nat Struct Biol 1998;5:793–802. [DOI] [PubMed] [Google Scholar]
- 14. Conibear E, Cleck JN, Stevens TH. Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t‐SNARE Tlg1p. Mol Biol Cell 2003;14:1610–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shestakova A, Suvorova E, Pavliv O, Khaidakova G, Lupashin V. Interaction of the conserved oligomeric Golgi complex with t‐SNARE Syntaxin5a/Sed5 enhances intra‐Golgi SNARE complex stability. J Cell Biol 2007;179:1179–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stroupe C, Collins KM, Fratti RA, Wickner W. Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J 2006;25:1579–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suvorova ES, Duden R, Lupashin VV. The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra‐Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J Cell Biol 2002;157:631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cai Y, Chin HF, Lazarova D, Menon S, Fu C, Cai H, Sclafani A, Rodgers DW, De La Cruz EM, Ferro‐Novick S, Reinisch KM. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane‐tethering complexes. Cell 2008;133:1202–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim YG, Sohn EJ, Seo J, Lee KJ, Lee HS, Hwang I, Whiteway M, Sacher M, Oh BH. Crystal structure of bet3 reveals a novel mechanism for Golgi localization of tethering factor TRAPP. Nat Struct Mol Biol 2005;12:38–45. [DOI] [PubMed] [Google Scholar]
- 20. Meier C, Aricescu AR, Assenberg R, Aplin RT, Gilbert RJ, Grimes JM, Stuart DI. The crystal structure of ORF‐9b, a lipid binding protein from the SARS coronavirus. Structure 2006;14:1157–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kummel D, Heinemann U, Veit M. Unique self‐palmitoylation activity of the transport protein particle component Bet3: a mechanism required for protein stability. Proc Natl Acad Sci U S A 2006;103:12701–12706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kummel D, Muller JJ, Roske Y, Misselwitz R, Bussow K, Heinemann U. The structure of the TRAPP subunit TPC6 suggests a model for a TRAPP subcomplex. EMBO Rep 2005;6:787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Turnbull AP, Kummel D, Prinz B, Holz C, Schultchen J, Lang C, Niesen FH, Hofmann KP, Delbruck H, Behlke J, Muller EC, Jarosch E, Sommer T, Heinemann U. Structure of palmitoylated BET3: insights into TRAPP complex assembly and membrane localization. EMBO J 2005;24:875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cai H, Yu S, Menon S, Cai Y, Lazarova D, Fu C, Reinisch K, Hay JC, Ferro‐Novick S. TRAPPI tethers COPII vesicles by binding the coat subunit Sec23. Nature 2007;445:941–944. [DOI] [PubMed] [Google Scholar]
- 25. Morozova N, Liang Y, Tokarev AA, Chen SH, Cox R, Andrejic J, Lipatova Z, Sciorra VA, Emr SD, Segev N. TRAPPII subunits are required for the specificity switch of a Ypt‐Rab GEF. Nat Cell Biol 2006;8:1263–1269. [DOI] [PubMed] [Google Scholar]
- 26. Segev N. Ypt and Rab GTPases: insight into functions through novel interactions. Curr Opin Cell Biol 2001;13:500–511. [DOI] [PubMed] [Google Scholar]
- 27. Lippe R, Horiuchi H, Runge A, Zerial M. Expression, purification, and characterization of Rab5 effector complex, rabaptin‐5/rabex‐5. Methods Enzymol 2001;329:132–145. [DOI] [PubMed] [Google Scholar]
- 28. Wickner W. Yeast vacuoles and membrane fusion pathways. EMBO J 2002;21:1241–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jedd G, Richardson C, Litt R, Segev N. The Ypt1 GTPase is essential for the first two steps of the yeast secretory pathway. J Cell Biol 1995;131:583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Segev N, Mulholland J, Botstein D. The yeast GTP‐binding YPT1 protein and a mammalian counterpart are associated with the secretion machinery. Cell 1988;52:915–924. [DOI] [PubMed] [Google Scholar]
- 31. Segev N. Mediation of the attachment or fusion step in vesicular transport by the GTP‐binding Ypt1 protein. Science 1991;252:1553–1556. [DOI] [PubMed] [Google Scholar]
- 32. Jones S, Richardson CJ, Litt RJ, Segev N. Identification of regulators for Ypt1 GTPase nucleotide cycling. Mol Biol Cell 1998;9:2819–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jones S, Litt RJ, Richardson CJ, Segev N. Requirement of nucleotide exchange factor for Ypt1 GTPase mediated protein transport. J Cell Biol 1995;130:1051–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jones S, Newman C, Liu F, Segev N. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol Biol Cell 2000;11:4403–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang W, Sacher M, Ferro‐Novick S. TRAPP stimulates guanine nucleotide exchange on Ypt1p. J Cell Biol 2000;151:289–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sacher M, Barrowman J, Wang W, Horecka J, Zhang Y, Pypaert M, Ferro‐Novick S. TRAPP I implicated in the specificity of tethering in ER‐to‐Golgi transport. Mol Cell 2001;7:433–442. [DOI] [PubMed] [Google Scholar]
- 37. Wurmser AE, Sato TK, Emr SD. New component of the vacuolar class C‐Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE‐dependent docking and fusion. J Cell Biol 2000;151:551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Walch‐Solimena C, Collins RN, Novick PJ. Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post‐Golgi vesicles. J Cell Biol 1997;137:1495–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hama H, Tall GG, Horazdovsky BF. Vps9p is a guanine nucleotide exchange factor involved in vesicle‐mediated vacuolar protein transport. J Biol Chem 1999;274:15284–15291. [DOI] [PubMed] [Google Scholar]
- 40. Siniossoglou S, Peak‐Chew SY, Pelham HR. Ric1p and Rgp1p form a complex that catalyses nucleotide exchange on Ypt6p. EMBO J 2000;19:4885–4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell 2007;129:865–877. [DOI] [PubMed] [Google Scholar]
- 42. Jedd G, Mulholland J, Segev N. Two new Ypt GTPases are required for exit from the yeast trans‐Golgi compartment. J Cell Biol 1997;137:563–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cai H, Zhang Y, Pypaert M, Walker L, Ferro‐Novick S. Mutants in trs120 disrupt traffic from the early endosome to the late Golgi. J Cell Biol 2005;171:823–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liang Y, Morozova N, Tokarev AA, Mulholland JW, Segev N. The role of Trs65 in the Ypt/Rab guanine nucleotide exchange factor function of the TRAPP II complex. Mol Biol Cell 2007;18:2533–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang W, Ferro‐Novick S. A Ypt32p exchange factor is a putative effector of Ypt1p. Mol Biol Cell 2002;13:3336–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yamamoto K, Jigami Y. Mutation of TRS130, which encodes a component of the TRAPP II complex, activates transcription of OCH1 in Saccharomyces cerevisiae. Curr Genet 2002;42:85–93. [DOI] [PubMed] [Google Scholar]
- 47. Zhang CJ, Bowzard JB, Greene M, Anido A, Stearns K, Kahn RA. Genetic interactions link ARF1, YPT31/32 and TRS130. Yeast 2002;19:1075–1086. [DOI] [PubMed] [Google Scholar]
- 48. Sciorra VA, Audhya A, Parsons AB, Segev N, Boone C, Emr SD. Synthetic genetic array analysis of the PtdIns 4‐kinase Pik1p identifies components in a Golgi‐specific Ypt31/rab‐GTPase signaling pathway. Mol Biol Cell 2005;16:776–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jiang Y, Scarpa A, Zhang L, Stone S, Feliciano E, Ferro‐Novick S. A high copy suppressor screen reveals genetic interactions between BET3 and a new gene. Evidence for a novel complex in ER‐to‐Golgi transport. Genetics 1998;149:833–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rossi G, Kolstad K, Stone S, Palluault F, Ferro‐Novick S. BET3 encodes a novel hydrophilic protein that acts in conjunction with yeast SNAREs. Mol Biol Cell 1995;6:1769–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 1994;77:895–907. [DOI] [PubMed] [Google Scholar]
- 52. Yoshihisa T, Barlowe C, Schekman R. Requirement for a GTPase‐activating protein in vesicle budding from the endoplasmic reticulum. Science 1993;259:1466–1468. [DOI] [PubMed] [Google Scholar]
- 53. Sato K, Nakano A. Dissection of COPII subunit‐cargo assembly and disassembly kinetics during Sar1p‐GTP hydrolysis. Nat Struct Mol Biol 2005;12:167–174. [DOI] [PubMed] [Google Scholar]
- 54. Mogelsvang S, Gomez‐Ospina N, Soderholm J, Glick BS, Staehelin LA. Tomographic evidence for continuous turnover of Golgi cisternae in Pichia pastoris. Mol Biol Cell 2003;14:2277–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Watson P, Forster R, Palmer KJ, Pepperkok R, Stephens DJ. Coupling of ER exit to microtubules through direct interaction of COPII with dynactin. Nat Cell Biol 2005;7:48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cao X, Ballew N, Barlowe C. Initial docking of ER‐derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J 1998;17:2156–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Richardson CJ, Jones S, Litt RJ, Segev N. GTP hydrolysis is not important for Ypt1 GTPase function in vesicular transport. Mol Cell Biol 1998;18:827–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lian JP, Ferro‐Novick S. Bos1p, an integral membrane protein of the endoplasmic reticulum to Golgi transport vesicles, is required for their fusion competence. Cell 1993;73:735–745. [DOI] [PubMed] [Google Scholar]
- 59. Allan BB, Moyer BD, Balch WE. Rab1 recruitment of p115 into a cis‐SNARE complex: programming budding COPII vesicles for fusion. Science 2000;289:444–448. [DOI] [PubMed] [Google Scholar]
- 60. Moyer BD, Allan BB, Balch WE. Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis–Golgi tethering. Traffic 2001;2:268–276. [DOI] [PubMed] [Google Scholar]
- 61. Chen SH, Chen S, Tokarev AA, Liu F, Jedd G, Segev N. Ypt31/32 GTPases and their novel F‐box effector protein Rcy1 regulate protein recycling. Mol Biol Cell 2005;16:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Conibear E, Stevens TH. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell 2000;11:305–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zolov SN, Lupashin VV. Cog3p depletion blocks vesicle‐mediated Golgi retrograde trafficking in HeLa cells. J Cell Biol 2005;168:747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Andag U, Neumann T, Schmitt HD. The coatomer‐interacting protein Dsl1p is required for Golgi‐to‐endoplasmic reticulum retrieval in yeast. J Biol Chem 2001;276:39150–39160. [DOI] [PubMed] [Google Scholar]
- 65. Gedeon AK, Colley A, Jamieson R, Thompson EM, Rogers J, Sillence D, Tiller GE, Mulley JC, Gecz J. Identification of the gene (SEDL) causing X‐linked spondyloepiphyseal dysplasia tarda. Nat Genet 1999;22:400–404. [DOI] [PubMed] [Google Scholar]
- 66. Ghosh AK, Steele R, Ray RB. Modulation of human luteinizing hormone beta gene transcription by MIP‐2A. J Biol Chem 2003;278:24033–24038. [DOI] [PubMed] [Google Scholar]
- 67. Hu WH, Pendergast JS, Mo XM, Brambilla R, Bracchi‐Ricard V, Li F, Walters WM, Blits B, He L, Schaal SM, Bethea JR. NIBP, a novel NIK and IKK(beta)‐binding protein that enhances NF‐(kappa)B activation. J Biol Chem 2005;280:29233–29241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Loh E, Peter F, Subramaniam VN, Hong W. Mammalian Bet3 functions as a cytosolic factor participating in transport from the ER to the Golgi apparatus. J Cell Sci 2005;118:1209–1222. [DOI] [PubMed] [Google Scholar]
- 69. Yamakawa K, Mitchell S, Hubert R, Chen XN, Colbern S, Huo YK, Gadomski C, Kim UJ, Korenberg JR. Isolation and characterization of a candidate gene for progressive myoclonus epilepsy on 21q22.3. Hum Mol Genet 1995;4:709–716. [DOI] [PubMed] [Google Scholar]
- 70. Sacher M, Ferro‐Novick S. Purification of TRAPP from Saccharomyces cerevisiae and identification of its mammalian counterpart. Methods Enzymol 2001;329:234–241. [DOI] [PubMed] [Google Scholar]
- 71. Yu S, Satoh A, Pypaert M, Mullen K, Hay JC, Ferro‐Novick S. mBet3p is required for homotypic COPII vesicle tethering in mammalian cells. J Cell Biol 2006;174:359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ghosh AK, Majumder M, Steele R, White RA, Ray RB. A novel 16‐kilodalton cellular protein physically interacts with and antagonizes the functional activity of c‐myc promoter‐binding protein 1. Mol Cell Biol 2001;21:655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fan L, Yu W, Zhu X. Interaction of Sedlin with chloride intracellular channel proteins. FEBS Lett 2003;540:77–80. [DOI] [PubMed] [Google Scholar]
- 74. Park JS, Lee ML, Jeong MS, Jin GE, Jang SB. The binding of human CLIC1 with SEDL and its characterization in vitro . Bull Korean Chem Soc 2007;28:574–580. [Google Scholar]
- 75. Gwynn B, Smith RS, Rowe LB, Taylor BA, Peters LL. A mouse TRAPP‐related protein is involved in pigmentation. Genomics 2006;88:196–203. [DOI] [PubMed] [Google Scholar]
- 76. Dell’Angelica EC. Melanosome biogenesis: shedding light on the origin of an obscure organelle. Trends Cell Biol 2003;13:503–506. [DOI] [PubMed] [Google Scholar]
- 77. Kaytor MD, Livingston DM. GSG1, a yeast gene required for sporulation. Yeast 1995;11:1147–1155. [DOI] [PubMed] [Google Scholar]
- 78. Meiling‐Wesse K, Epple UD, Krick R, Barth H, Appelles A, Voss C, Eskelinen EL, Thumm M. Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J Biol Chem 2005;280:33669–33678. [DOI] [PubMed] [Google Scholar]
- 79. Nazarko TY, Huang J, Nicaud JM, Klionsky DJ, Sibirny AA. Trs85 is required for macroautophagy, pexophagy and cytoplasm to vacuole targeting in Yarrowia lipolytica and Saccharomyces cerevisiae. Autophagy 2005;1:37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cox R, Chen SH, Yoo E, Segev N. Conservation of the TRAPPII‐specific subunits of a Ypt/Rab exchanger complex. BMC Evol Biol 2007;7:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H et al Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 2002;415:141–147. [DOI] [PubMed] [Google Scholar]
- 82. Koumandou VL, Dacks JB, Coulson RM, Field MC. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol Biol 2007;7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chiari R, Foury F, De PE, Baurain JF, Thonnard J, Coulie PG. Two antigens recognized by autologous cytolytic T lymphocytes on a melanoma result from a single point mutation in an essential housekeeping gene. Cancer Res 1999;59:5785–5792. [PubMed] [Google Scholar]
- 84. Shaw MA, Brunetti‐Pierri N, Kadasi L, Kovacova V, Van Maldergem L, De Brasi D, Salerno M, Gecz J. Identification of three novel SEDL mutations, including mutation in the rare, non‐canonical splice site of exon 4. Clin Genet 2003;64:235–242. [DOI] [PubMed] [Google Scholar]
- 85. Tiller GE, Hannig VL, Dozier D, Carrel L, Trevarthen KC, Wilcox WR, Mundlos S, Haines JL, Gedeon AK, Gecz J. A recurrent RNA‐splicing mutation in the SEDL gene causes X‐linked spondyloepiphyseal dysplasia tarda. Am J Hum Genet 2001;68:1398–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sacher M. Membrane traffic fuses with cartilage development. FEBS Lett 2003;550:1–4. [DOI] [PubMed] [Google Scholar]
- 87. Gecz J, Hillman MA, Gedeon AK, Cox TC, Baker E, Mulley JC. Gene structure and expression study of the SEDL gene for spondyloepiphyseal dysplasia tarda. Genomics 2000;69:242–251. [DOI] [PubMed] [Google Scholar]
- 88. Schlenker O, Hendricks A, Sinning I, Wild K. The structure of the mammalian signal recognition particle (SRP) receptor as prototype for the interaction of small GTPases with Longin domains. J Biol Chem 2006;281:8898–8906. [DOI] [PubMed] [Google Scholar]
- 89. Gecz J, Shaw MA, Bellon JR, De Barros LM. Human wild‐type SEDL protein functionally complements yeast Trs20p but some naturally occurring SEDL mutants do not. Gene 2003;320:137–144. [DOI] [PubMed] [Google Scholar]


