Abstract
Parotid Secretory Protein (PSP/SPLUNC2) is expressed in human salivary glands and saliva. The protein exists as an N-glycosylated and non-glycosylated form and both appear to induce agglutination of bacteria, a major antibacterial function for salivary proteins. Both forms of PSP/SPLUNC2 bind lipopolysaccharide suggesting that the protein may also play an anti-inflammatory role. Based on the predicted structure of PSP/SPLUNC2 and the location of known antibacterial and anti-inflammatory peptides in bactericidal/permeability-increasing protein and lipopolysaccharide-binding protein, we designed the synthetic peptides GL13NH2 and GL13K that capture these proposed functions of PSP/SPLUNC2. GL13NH3 agglutinates bacteria leading to increased clearance by macrophages and reduced spread of infection in a plant model. GL13K kills bacteria with a minimal inhibitory concentration of 5–10 µg/ml, kills bacteria in biofilm and retains activity in 150 mM NaCl and 50% saliva. Both peptides block endotoxin action but only GL13K appears to bind endotoxin. The peptides do not cause hemolysis, hemagglutination in serum, inhibit mammalian cell proliferation or induce an inflammatory response in macrophages. These results suggest that the GL13NH2 and the modified peptide GL13K capture the biological activity of PSP/SPLUNC2 and can serve as lead compounds for the development of novel antimicrobial and anti-inflammatory peptides.
Keywords: Parotid secretory protein, antimicrobial, surfactant, agglutination, lipid-binding, lipopolysaccharide, endotoxin, C20orf70
Introduction
Parotid Secretory Protein (PSP)/short palate, lung, and nasal epithelium clone 2 (SPLUNC2) (C20orf70) is the predominant member of the palate, lung and nasal epithelium clone (PLUNC) family in saliva. PSP was originally identified in the mouse parotid gland and it has long been recognized as a major secretory protein in rodent parotid glands [1–3]. PSP is also expressed in the mouse lacrimal gland [4] but is not among 54 proteins identified in human tears [5]. Hamster PSP is expressed in salivary glands and the protein was also found in high density lipoprotein, which could indicate a functional overlap between PSP and lipid transport proteins, including cholesteryl ester transport protein (CETP) and phospholipid transfer protein (PTLP) [6]. Horse PSP is an abundant 28 kD protein that is expressed in salivary glands and sweat [7]. It is related to latherin, which is also expressed in these fluids [8, 9]. A group of PSP-related proteins, BSP30s [10, 11], are highly expressed in bovine saliva, in particular in cattle with low susceptibility to pasture bloat [12]. If confirmed, this association could point to an antibacterial or surfactant function of BSP30s in the rumen. PSP is also expressed in pig (Sus scrofa) salivary glands [13]. The high expression of PSP in salivary glands has prompted use of its promoter to specifically target transgenic phytase expression to saliva to reduce phosphorus pollution from animal manure [14–16]. PSP-related proteins have also been predicted in dog (Canis familiaris); chimpanzee (Pan troglodytes) (NCBI ID: XP_001156872); orangutan (Pongo abelii) (XP_002830248); Rhesus macaque (Macaca mulatta) (XP_002830248); marmoset (Callithrix jacchus) (XP_002747268); giant panda (Ailuropoda melanoleuca) (XP_002918340). This list suggests that PSP/SPLUNC2 may be generally required for salivary function, independent of a specific diet.
Characterization of human PSP/SPLUNC2
Human PSP/SPLUNC2 is expressed in salivary glands, saliva and gingival epithelial cells [17, 18]. In the latter cell type, PSP/SPLUNC2 expression is regulated by the Gram-negative bacteria Porphyromonas gingivalis and the pro-inflammatory cytokine TNFα [18]. In the human parotid gland PSP/SPLUNC2 is expressed in both acinar and ductal epithelial cells [17]. PSP/SPLUNC2 has been identified in glandular saliva from the parotid and submandibular/sublingual glands. Unlike horse, cow and rodent PSP/BSP30, the human protein is not a major band detected by SDS-PAGE of whole saliva (unpublished observation) but the protein appears in the top 30 of over 900 salivary proteins based on the number of distinct peptides identified by proteomic analysis [19]. The functional significance of this difference between species, if any, is not known.
Human PSP/SPLUNC2 is a 249 amino acid protein. The protein contains two N-glycosylation sites located at Asn(124) and Asn(132) [20]. The protein is typically found by immunoblotting of saliva as two major distinct bands likely representing the N-glycosylated and non-glycosylated forms. Consistent with this, we found that PSP/SPLUNC2 expressed in mammalian cells corresponds to the upper band seen in saliva while PSP/SPLUNC2 expressed in E. coli corresponds to the lower band [17]. In addition, the upper bands are collapsed to a lower molecular weight band upon treatment with endoglycosidase F [21].
Based on sequence prediction analyses, databases currently identify amino acid residues 1–18 as the putative signal sequence of PSP/SPLUNC2 (e.g. UniProt ID Q96DR5). However, trypsin digests of proteins secreted in saliva (which would lack the signal peptide) contain PSP/SPLUNC2 peptides with one or two additional N-terminal residues, including T.SESLLDNLGNDLSNVVDKLEPVLHEGLETVDNTLK.G [19]. Based on this analysis, we propose that the signal peptide of PSP/SPLUNC2 consists of residues 1–16 and the mature protein consists of residues 17–249.
Functional models for PSP/SPLUNC2
Salivary proteins typically function in wetting and digestion of food (e.g. mucins, amylase), protection and remineralization of tooth surfaces (e.g. mucin, proline-rich proteins) or host defense of the oral cavity (e.g. lysozyme, defensins) [22]. PSP/SPLUNC2 is related to the BPI/LBP/CETP family of mammalian lipid binding proteins that includes bactericidal/permeability increasing protein (BPI), lipopolysaccharide-binding protein (LBP), cholesteryl ester transport protein (CETP) and phospholipid transport protein (PLTP) [23]. Indeed, PSP/SPLUNC2 and its related proteins contain conserved Cys residues that form a disulfide bridge in BPI. The disulfide bridge has not been verified in PSP/SPLUNC2, but a PSP/SPLUNC2 mutant lacking the Cys residues is not secreted, suggesting that its structure is significantly altered (Geetha and Gorr, unpublished observation). Aside from the conserved Cys residues, the sequence identity of PSP/SPLUNC2/SPLUNC2 and BPI is low (22%), consistent with observations for other proteins related to the BPI family [24]. Based on the known functions of salivary proteins, the similarity of PSP/SPLUNC2 to BPI/LBP/CETP proteins and the mucosal localization of the PLUNC family, we focused our functional studies on host-defense functions of PSP/SPLUNC2. This is supported by the findings that mouse PSP binds bacteria [4] and that PLUNC exhibits anti-mycoplasma activity, anti-pseudomonal activity and binds lipopolysaccharide (LPS) [25, 26].
Antibacterial function of PSP/SPLUNC2
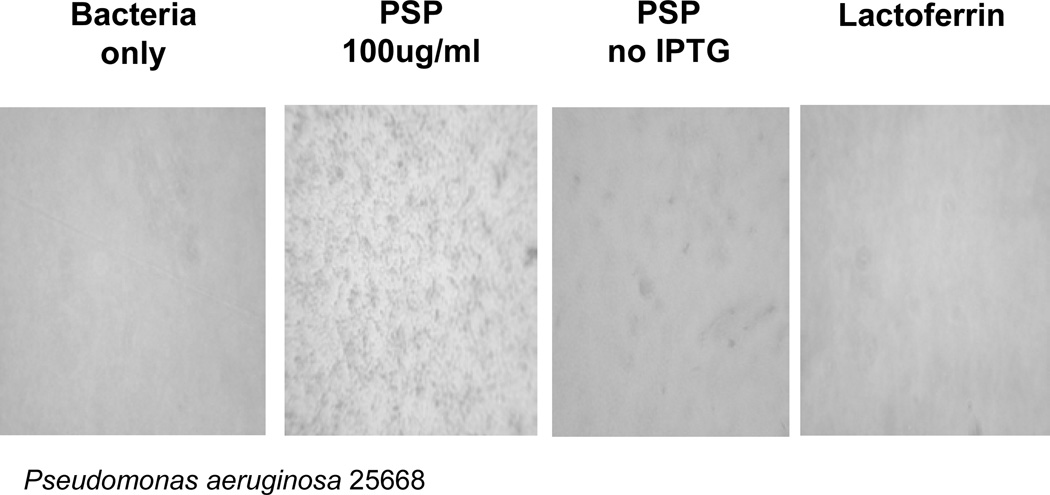
We initially expressed PSP/SPLUNC2 in the rat cell line GH4C1 and used the secretion medium to test the effect on the number of colony forming units. Secretion media that contained PSP/SPLUNC2 reduced the number of colonies of Pseudomonas aeruginosa PAO1 [17]. We originally interpreted this reduction to indicate a bactericidal activity of PSP/SPLUNC2 but subsequent studies with PSP-peptides [27] indicated that bacterial agglutination also would result in a reduction in colony forming units. To test if PSP/SPLUNC2 causes bacterial agglutination, the His-tagged protein was expressed in E. coli and inclusion bodies collected and solubilized (T. Wheeler, AgResearch, New Zealand; personal communication). The crude PSP/SPLUNC2 samples contained about 45% PSP/SPLUNC2 as judged from SDS-PAGE and Coomassie blue staining. This fraction was incubated with P. aeruginosa and bacterial agglutination was observed by light microscopy (Figure 1). Only samples from PSP/SPLUNC2 producing cultures that had been induced with IPTG caused agglutination. These results support the proposal that PSP/SPLUNC2 acts as a salivary agglutinin similar to DMBT1 [27].
Figure 1. Agglutination of P. aeruginosa by recombinant PSP/SPLUNC2.
P. aeruginosa 2566 were incubated with PSP/SPLUNC2 expressed in induced (PSP 100 µg/ml) or un-induced culture (PSP no IPTG). Bacteria only and Lactoferrin were used as controls. Bacterial agglutination was identified by light microscopy of the culture dish.
Antibacterial function of PSP-peptides
Based on the predicted similarity of PSP/SPLUNC2 and the BPI/LBP proteins as well as the known location of biologically active peptides in the sequences of BPI and LBP [28], we designed PSP/SPLUNC2 peptides with predicted antibacterial and anti-inflammatory activity. Indeed, peptides were identified that inhibit the action of LPS [29] and cause agglutination of both Gram negative (P. aeruginosa; Aggregatibacter actinomycetemcomitans) and Gram positive (Streptococcus gordonii) bacteria [27]. Agglutination may play a biological role since the agglutinating peptide GL13NH2 (PSP 141–153) limited the spread of P. aeruginosa in a lettuce leaf infection model and increased the uptake of P. aeruginosa by macrophages [27].
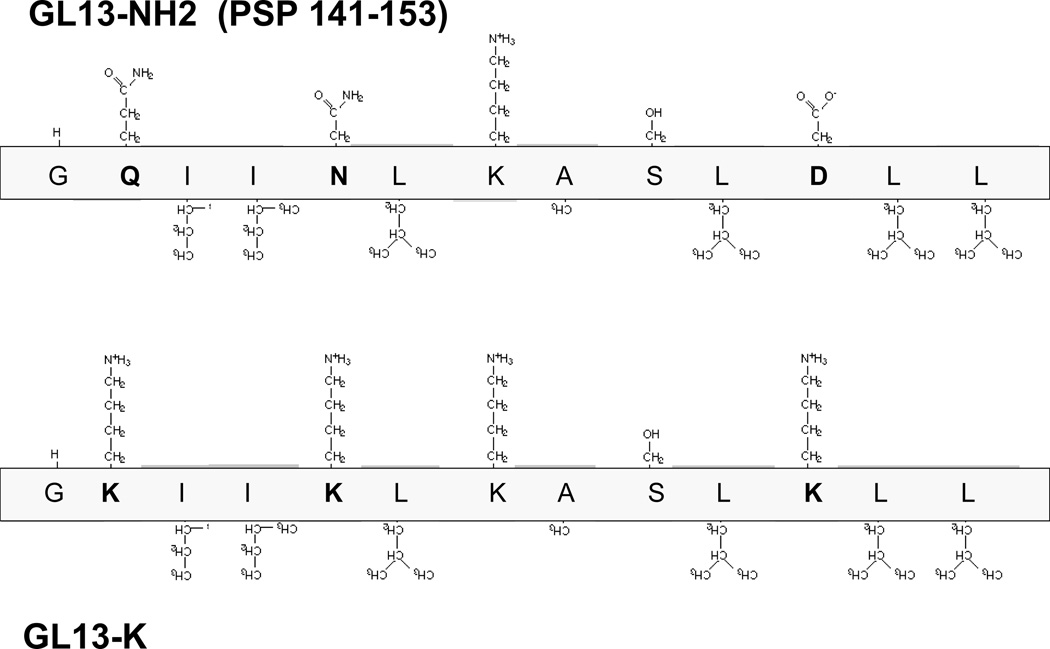
GL13NH2 did not exhibit bactericidal activity. To achieve this, the peptide was modified to increase the overall positive charge by substituting three amino acids for Lysine residues (Figure 2). This modified peptide (GL13K) did not exhibit agglutinating activity but, instead, exhibited bactericidal activity in a 2 h kill assay performed in 10 mM sodium phosphate [e.g.30]. GL13K and the control peptide polymyxin B effectively killed the Gram negative bacteria P. aeruginosa, E. coli and the Gram positive bacteria S. gordonii, whereas other PSP/SPLUNC2 peptides had no effect on bacterial survival. The bactericidal activity of GL13K was not affected by the addition of 150 mM NaCl or 50% saliva. The minimal inhibitory concentration of GL13K, as determined by microtiter broth dilution [31, 32], was 5–10 µg/ml against P. aeruginosa and E.coli.
Figure 2. Sequence and side chain structure of the PSP peptides GL13NH2 (top) and GL13K (bottom).
The amino acids are oriented with hydrophilic side chains facing up and hydrophobic side chain facing down. Residues that differ between the two peptides are shown in bold.
To test if GL13K was effective against bacteria grown in a biofilm, P. aeruginosa were cultured for 24 hours in 96-well PVC plates, which were washed to remove planktonic bacteria. The wells were treated with GL13K or control peptides and surviving attached bacteria quantitated by an ATP viability assay. GL13K and polymyxin B reduced viable attached bacteria by over 90%. Other PSP-peptides or buffer alone had no effect on viability.
Lipopolysaccharide–binding activity of PSP/SPLUNC2
PLUNC has been reported to bind LPS [25], as expected from its structural similarity to LBP, whereas LPS-binding was not detected for BSP30 [11]. To determine if PSP/SPLUNC2 binds LPS, whole saliva samples were incubated with LPS-beads, which were then washed and analyzed by immunoblotting. Both the glycosylated and unglycosylated forms of PSP/SPLUNC2 were captured by LPS and could be eluted with a non-ionic detergent, suggesting a hydrophobic interaction. Moreover, binding of PSP/SPLUNC2 to LPS was inhibited by GL13NH2 suggesting that this peptide interferes with the binding site. GL13K was not tested in this assay but the two peptides (GL13NH2 and GL13K) inhibited the LPS-stimulated secretion of TNFα from macrophages by 60% and 80% respectively.
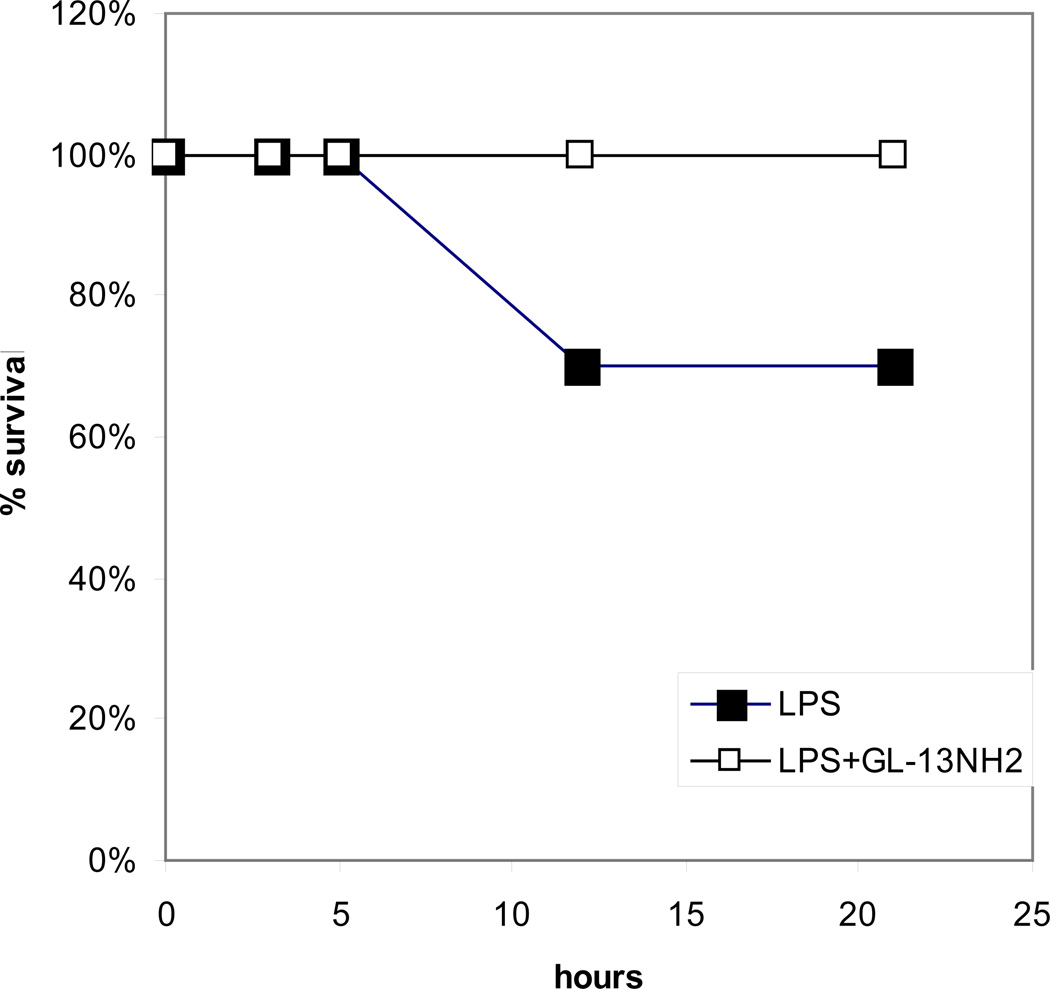
In preliminary experiments, we have tested the activity of GL-13NH2 in a D-galactosamine-sensitized mouse sepsis model. C57BL/6 mice were sensitized with D-galactosamine and injected i.p. with P. aeruginosa LPS. Death typically occurred in 8–10 hours in this model. LPS was injected alone or in combination with GL13NH2 (Figure 3). GL13NH2 increased survival in this model from 70% to 100%.
Figure 3. Survival in a mouse sepsis model.
C57BL/6 mice were sensitized with D-galactosamine and injected with 100 ng/mouse (50µg/kg) of P. aeruginosa LPS that had been preincubated for 1h RT with or without 100 µg/mouse of GL13NH2. Controls received LPS and 0.9% saline. The mice were observed at the times indicated and % survival calculated. The results are from two independent experiments.
Toxicity studies
Polymyxin is currently in clinical use for ophthalmic and skin infections. It was disused for several years due to its high toxicity [33]. Thus, there is a need for novel, less toxic antibiotics. To test the toxicity of PSP-peptides we evaluated the lysis of sheep red blood cells, hemagglutination, survival and proliferation of mammalian cells and the inflammatory response in macrophages (TNFα secretion). While GL13NH2 caused hemagglutination in the absence of serum, none of the other assays indicated toxicity of these peptides. Importantly, all mice that received GL13NH2 survived (Figure 3). In addition, we maintained Caenorhabditis. elegans on P. aeruginosa with or without GL13NH2 and observed no differences in viability or hatching of young worms.
Summary
The data reviewed here strongly support a host-defense function for PSP/SPLUNC2. Indeed, we have found evidence that both the intact protein and peptides derived from the protein sequence exhibit antibacterial activity and anti-inflammatory activity. Thus, these data support the biological functions that were predicted by the proposed structural similarity of PLUNC proteins and BPI/LBP. These results are also consistent with the mucosal expression of PLUNC proteins. It is important to consider, however, that the functional predictions are based on computer modeling of the PSP/SPLUNC2 sequence and that related proteins are involved in lipid binding (CETP, PLTP), juvenile hormone binding, odorant binding and olfaction. In addition, PLUNC [34] and latherin [9] display surfactant activity that may contribute to the wetting of mucosal and skin surfaces. Indeed, we have recently reported that the PSP-peptide GL13NH2 is more closely related to biologically active peptides from surfactant protein and salivary agglutinin (DMBT1) than BPI [27]. Consistent with this, we recently found that surfaces coated with GL13K exhibit an increased advancing water contact angle, as would be expected of a surfactant [35]. These, currently disparate, findings add to the facets of an interesting protein that is expressed in varying abundance in the saliva of different animals.
Acknowledgements
We thank Dr. Thomas Wheeler, AgResearch, New Zealand for sharing unpublished data. The research described here was supported by U.S. Public Health Service grants [1R01DE012205] and [1R01DE017989] from the National Institute for Dental and Craniofacial Research. Additional support from the University of Louisville and University of Minnesota Schools of Dentistry is gratefully acknowledged.
Abbreviations
- BPI
bactericidal/permeability-increasing protein
- CETP
cholesteryl ester transfer protein
- LBP
lipopolysaccharide-binding protein
- LPS
lipopolysaccharide
- PLTP
phospholipid transfer protein
- PLUNC
palate, lung, and nasal epithelium clone
- PSP
Parotid Secretory Protein
- SPLUNC2
short palate, lung, and nasal epithelium clone 2
BIBLIOGRAPHY
- 1.Mirels L, Ball WD. Neonatal Rat Submandibular-Gland Protein Smg-a and Parotid Secretory Protein Are Alternatively Regulated Members of a Salivary Protein Multigene Family. Journal of Biological Chemistry. 1992;267:2679–2687. [PubMed] [Google Scholar]
- 2.Madsen HO, Hjorth JP. Molecular cloning of mouse PSP mRNA. Nucleic Acids Res. 1985;13:1–13. doi: 10.1093/nar/13.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venkatesh SG, Cowley DJ, Gorr S-U. Differential aggregation properties of secretory proteins that are stored in exocrine secretory granules of the pancreas and parotid glands. Am J Physiol Cell Physiol. 2004;286:C365–C371. doi: 10.1152/ajpcell.00338.2003. [DOI] [PubMed] [Google Scholar]
- 4.Robinson CP, Bounous DI, Alford CE, Nguyen KH, Nanni JM, Peck AB, Humphreys-Beher MG. PSP expression in murine lacrimal glands and function as a bacteria binding protein in exocrine secretions. Am J Physiol. 1997;272:G863–G871. doi: 10.1152/ajpgi.1997.272.4.G863. [DOI] [PubMed] [Google Scholar]
- 5.Li N, Wang N, Zheng J, Liu XM, Lever OW, Erickson PM, Li L. Characterization of Human Tear Proteome Using Multiple Proteomic Analysis Techniques. J. Proteome Res. 2005;4:2052–2061. doi: 10.1021/pr0501970. [DOI] [PubMed] [Google Scholar]
- 6.Khovidhunkit W, Hachem JP, Medzihradszky KF, Duchateau PN, Shigenaga JK, Moser AH, Movsesyan I, Naya-Vigne J, Kane JP, Feingold KR, Grunfeld C. Parotid secretory protein (PSP) is an HDL-associated protein with anticandidal activity. Am J Physiol Regul Integr Comp Physiol. 2005;288:R1306–R1315. doi: 10.1152/ajpregu.00007.2004. [DOI] [PubMed] [Google Scholar]
- 7.D'Innocenzo B, Salzano AM, D'Ambrosio C, Gazzano A, Niccolini A, Sorce C, Dani FR, Scaloni A, Pelosi P. Secretory Proteins as Potential Semiochemical Carriers in the Horse. Biochemistry. 2006;45:13418–13428. doi: 10.1021/bi061409p. [DOI] [PubMed] [Google Scholar]
- 8.Beeley JG, Eason R, Snow DH. Isolation and characterization of latherin, a surface-active protein from horse sweat. Biochem. J. 1986;235:645–650. doi: 10.1042/bj2350645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDonald RE, Fleming RI, Beeley JG, Bovell DL, Lu JR, Zhao X, Cooper A, Kennedy MW. Latherin: a surfactant protein of horse sweat and saliva. PloS one. 2009;4:e5726. doi: 10.1371/journal.pone.0005726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wheeler T, Hood K, Maqbool N, McEwan J, Bingle C, Zhao S. Expansion of the Bactericidal/permeability increasing-like (BPI-like) protein locus in cattle. BMC Genomics. 2007;8:75. doi: 10.1186/1471-2164-8-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haigh B, Hood K, Broadhurst M, Medele S, Callaghan M, Smolenski G, Dines M, Wheeler T. The bovine salivary proteins BSP30a and BSP30b are independently expressed BPI-like proteins with anti-Pseudomonas activity. Mol Immunol. 2008;45:1944–1951. doi: 10.1016/j.molimm.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 12.Rajan GH, Morris CA, Carruthers VR, Wilkins RJ, Wheeler TT. The relative abundance of a salivary protein, bSP30, is correlated with susceptibility to bloat in cattle herds selected for high or low bloat susceptibility. Anim Genet. 1996;27:407–414. doi: 10.1111/j.1365-2052.1996.tb00507.x. [DOI] [PubMed] [Google Scholar]
- 13.Yin HF, Zhao ZH, Fan BL, Liu ZL, Lu W, Liu YF, Li N. cDNA cloning, genomic structure, chromosomal mapping, and expression analysis of parotid secretory protein in pig. Genomics. 2004;83:9–18. doi: 10.1016/s0888-7543(03)00125-3. [DOI] [PubMed] [Google Scholar]
- 14.Golovan SP, Hayes MA, Phillips JP, Forsberg CW. Transgenic mice expressing bacterial phytase as a model for phosphorus pollution control. Nat Biotechnol. 2001;19:429–433. doi: 10.1038/88091. [DOI] [PubMed] [Google Scholar]
- 15.Golovan SP, Meidinger RG, Ajakaiye A, Cottrill M, Wiederkehr MZ, Barney DJ, Plante C, Pollard JW, Fan MZ, Hayes MA, Laursen J, Hjorth JP, Hacker RR, Phillips JP, Forsberg CW. Pigs expressing salivary phytase produce low-phosphorus manure. Nat Biotechnol. 2001;19:741–745. doi: 10.1038/90788. [DOI] [PubMed] [Google Scholar]
- 16.Yin HF, Fan BL, Yang B, Liu YF, Luo J, Tian XH, Li N. Cloning of pig parotid secretory protein gene upstream promoter and the establishment of a transgenic mouse model expressing bacterial phytase for agricultural phosphorus pollution control. J. Anim Sci. 2006;84:513–519. doi: 10.2527/2006.843513x. [DOI] [PubMed] [Google Scholar]
- 17.Geetha C, Venkatesh SG, Fasciotto Dunn BH, Gorr S-U. Expression and anti-bacterial activity of human parotid secretory protein (PSP) Biochem. Soc. Trans. 2003;31:815–818. doi: 10.1042/bst0310815. [DOI] [PubMed] [Google Scholar]
- 18.Shiba H, Venkatesh SG, Gorr SU, Barbieri G, Kurihara H, Kinane DF. Parotid secretory protein is expressed and inducible in human gingival keratinocytes. J Periodontal Res. 2005;40:153–157. doi: 10.1111/j.1600-0765.2005.00781.x. [DOI] [PubMed] [Google Scholar]
- 19.Denny P, Hagen FK, Hardt M, Liao L, Yan W, Arellanno M, Bassilian S, Bedi GS, Boontheung P, Cociorva D, Delahunty CM, Denny T, Dunsmore J, Faull KF, Gilligan J, Gonzalez-Begne M, Halgand F, Hall SC, Han X, Henson B, Hewel J, Hu S, Jeffrey S, Jiang J, Loo JA, Ogorzalek Loo RR, Malamud D, Melvin JE, Miroshnychenko O, Navazesh M, Niles R, Park SK, Prakobphol A, Ramachandran P, Richert M, Robinson S, Sondej M, Souda P, Sullivan MA, Takashima J, Than S, Wang J, Whitelegge JP, Witkowska HE, Wolinsky L, Xie Y, Xu T, Yu W, Ytterberg J, Wong DT, Yates JR, Fisher SJ. The Proteomes of Human Parotid and Submandibular/Sublingual Gland Salivas Collected as the Ductal Secretions. J. Proteome Res. 2008;7:1994–2006. doi: 10.1021/pr700764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramachandran P, Boontheung P, Xie Y, Sondej M, Wong DT, Loo JA. Identification of N-Linked Glycoproteins in Human Saliva by Glycoprotein Capture and Mass Spectrometry. J. Proteome Res. 2006;5:1493–1503. doi: 10.1021/pr050492k. [DOI] [PubMed] [Google Scholar]
- 21.Bingle L, Barnes F, Lunn H, Musa M, Webster S, Douglas C, Cross S, High A, Bingle C. Characterisation and expression of SPLUNC2, the human orthologue of rodent parotid secretory protein. Histochem. Cell Biol. 2009;132:339–349. doi: 10.1007/s00418-009-0610-4. [DOI] [PubMed] [Google Scholar]
- 22.Van Nieuw Amerongen A, Bolscher JG, Veerman EC. Salivary proteins: protective and diagnostic value in cariology? Caries Res. 2004;38:247–253. doi: 10.1159/000077762. [DOI] [PubMed] [Google Scholar]
- 23.Bingle CD, Gorr S-U. Host defense in oral and airway epithelia: chromosome 20 contributes a new protein family. Intl. J. Biochem. Cell Biol. 2004;36:2144–2152. doi: 10.1016/j.biocel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 24.Beamer LJ, Fischer D, Eisenberg D. Detecting distant relatives of mammalian LPS-binding and lipid transport proteins. Protein Sci. 1998;7:1643–1646. doi: 10.1002/pro.5560070721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghafouri B, Kihlstrom E, Tagesson C, Lindahl M. PLUNC in human nasal lavage fluid: multiple isoforms that bind to lipopolysaccharide. Biochim Biophys Acta. 2004;1699:57–63. doi: 10.1016/j.bbapap.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Zhou HD, Li XL, Li GY, Zhou M, Liu HY, Yang YX, Deng T, Ma J, Sheng SR. Effect of SPLUNC1 protein on the Pseudomonas aeruginosa and Epstein-Barr virus. Mol Cell Biochem. 2008;309:191–197. doi: 10.1007/s11010-007-9659-3. [DOI] [PubMed] [Google Scholar]
- 27.Gorr S-U, Sotsky JB, Shelar AP, Demuth DR. Design of bacteria-agglutinating peptides derived from Parotid Secretory Protein, a member of the Bactericidal/Permeability Increasing-like protein family. Peptides. 2008;29:18–27. doi: 10.1016/j.peptides.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 28.Dankesreiter S, Hoess A, Schneider-Mergener J, Wagner H, Miethke T. Synthetic endotoxin-binding peptides block endotoxin-triggered TNF-{alpha} production by macrophages in vitro and in vivo and prevent endotoxin-mediated toxic shock. J Immunol. 2000;164:4804–4811. doi: 10.4049/jimmunol.164.9.4804. [DOI] [PubMed] [Google Scholar]
- 29.Geetha C, Venkatesh SG, Bingle L, Bingle CD, Gorr SU. Design and Validation of Anti-inflammatory Peptides from Human Parotid Secretory Protein. J Dent Res. 2005;84:149–153. doi: 10.1177/154405910508400208. [DOI] [PubMed] [Google Scholar]
- 30.Haversen LA, Engberg I, Baltzer L, Dolphin G, Hanson LA, Mattsby-Baltzer I. Human Lactoferrin and Peptides Derived from a Surface-Exposed Helical Region Reduce Experimental Escherichia coli Urinary Tract Infection in Mice. Infect. Immun. 2000;68:5816–5823. doi: 10.1128/iai.68.10.5816-5823.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amsterdam D. Susceptibility testing of antimicrobials in liquid media. In: Lorian V, editor. Antibiotics in Laboratory Medicine. Baltimore, MD: Williams and Wilkins; 1996. pp. 52–111. [Google Scholar]
- 32.Hancock REW. Hancock Laboratory Methods. British Columbia, Canada: Department of Microbiology and Immunology, University of British Columbia; 1999. Modified MIC Method for Cationic Antimicrobial Peptides. http://www.cmdr.ubc.ca/bobh/methods.php. [Google Scholar]
- 33.Lim LM, Ly N, Anderson D, Yang JC, Macander L, Jarkowski A, 3rd, Forrest A, Bulitta JB, Tsuji BT. Resurgence of colistin: a review of resistance, toxicity, pharmacodynamics, and dosing. Pharmacotherapy. 2010;30:1279–1291. doi: 10.1592/phco.30.12.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gakhar L, Bartlett JA, Penterman J, Mizrachi D, Singh PK, Mallampalli RK, Ramaswamy S, McCray PB., Jr PLUNC is a novel airway surfactant protein with anti-biofilm activity. PloS one. 2010;5:e9098. doi: 10.1371/journal.pone.0009098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holmberg K, Hegde R, Abdolhosseini M, Gorr S-U, Aparicio CJ. San Diego, CA: American Association for Dental Research; 2011. Antimicrobial-Peptide Biofunctionalized Titanium for Dental Implants. [Google Scholar]