Fig. 2.
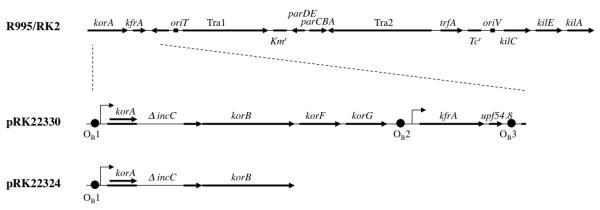
Linear schematic of key plasmids used to study the incompatibility properties of the incC2 mutants. The IncPα backbone for plasmids RK2 and R995 is shown at the top with landmark genetic determinants for reference. The primary difference between the IncPα plasmids, R995 and RK2, is transposon Tn1 in the kilC operon, which is present only in RK2 and is excluded from this schematic. The bold horizontal arrows in the R995/RK2 schematic refer to genetic loci or operons; in the pRK22330 and pRK22324 schematics, the arrows indicate genes. oriV and oriT refer to the plasmid origin of replication and the origin of conjugal transfer, respectively. Kmr denotes the gene for kanamycin resistance; Tcr denotes the tetracycline resistance locus. The angled arrows indicate promoters. The black circles, referred to as OB1, OB2 and OB3, indicate binding sites for KorB. The korA gene lies within the incC coding sequence, but in a different reading frame. ΔincC refers to a 465-bp in-frame deletion within incC (see text for details). The incC deletion present on R995ΔincC is the same as the one depicted here in the schematics of pRK22330 and pRK22324.
