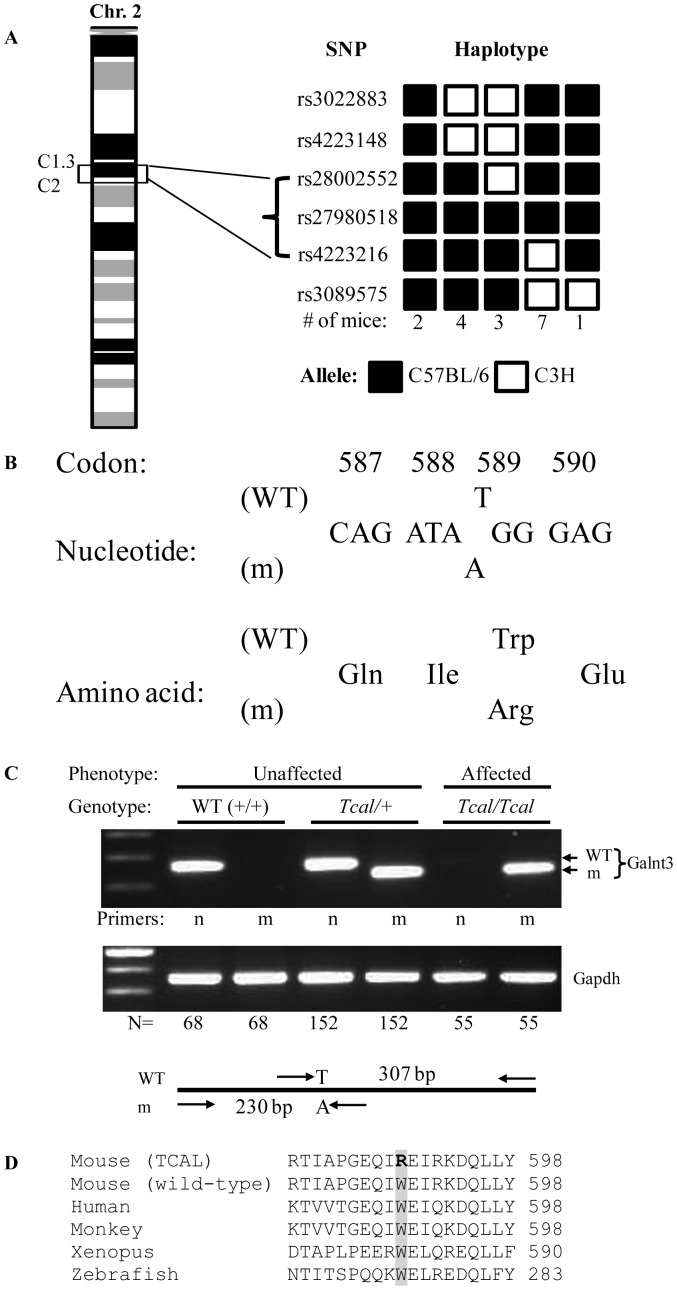
Figure 3. Mapping of Tcal locus and identification of Galnt3 mutation.
(A)The Tcal locus, which originated in a C57BL/6 ENU-mutagenised male and is hence inherited with the C57BL/6 alleles, was mapped to a 8.47 Mb region flanked by the SNPs rs28002552 and rs4223216 on chromosome 2C1.3–C2. This region contained 95 genes which included the Galnt3 gene. (B) DNA sequence analysis of Galnt3 identified a T to A transversion in codon 589, such that the wild type (WT) sequence, TGG which encodes an evolutionarily conserved tryptophan (Trp) residue was altered to the mutant (m) sequence, AGG which encodes an arginine (Arg) residue. (C) Amplification refractory mutation system (ARMS) PCR was used to confirm the presence of the mutation by designing primers (n, normal (WT) and m, mutant) that yielded 307 bp WT and 230 bp mutant PCR products, respectively. PCR amplification of Gapdh was used as a control for the presence of DNA. N = numbers of mice with each genotype. (D) Protein sequence alignment (CLUSTALW) of Galnt3 from 5 species revealed that the Trp (W) residue is evolutionarily conserved in the Galnt3 orthologues of mouse, human, monkey, xenopus and zebrafish.