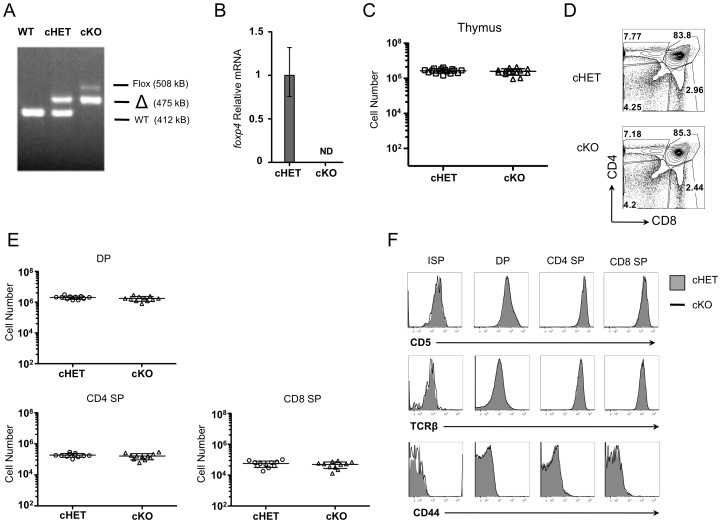
Figure 2. Deletion of Foxp4 at the DP stage does not alter thymocyte development.
A) DNA isolated from cHET and cKO thymocytes was amplified using primers to detect wild-type, floxed, and deleted allelic sequences. Band sizes and identities are indicated. Representative of ten experiments. B) RNA isolated from WT (C57BL/6) and cKO thymocytes was assessed for Foxp4 by real time PCR. ND = not detectable. Representative of three experiments. C) Total cellularity was assessed in both cHET and cKO thymi from four-week-old littermates. Each point represents a single mouse. Mean and standard deviation are indicated. D) Thymocytes were stained for CD4 and CD8 and analyzed by polychromatic flow. Contour plots shown are previously gated on live, singlet-gated cells, negative for myeloid lineage markers (CD11b, CD11C, CD19, B220). Gated frequencies are indicated. Representative of twelve cHET and twelve cKO mice. E) Absolute numbers of thymocyte populations were determined. Each point represents an individual mouse. F) Thymocytes were stained with CD4, CD8, CD5 and HSA. Cells are gated on populations as indicated at the top of each column and assessed for CD5, TCRβ, and CD44 expression. Solid histograms are from cHET and bold lines are from cKO mice. Representative of 12 cHET and 12 cKO mice.