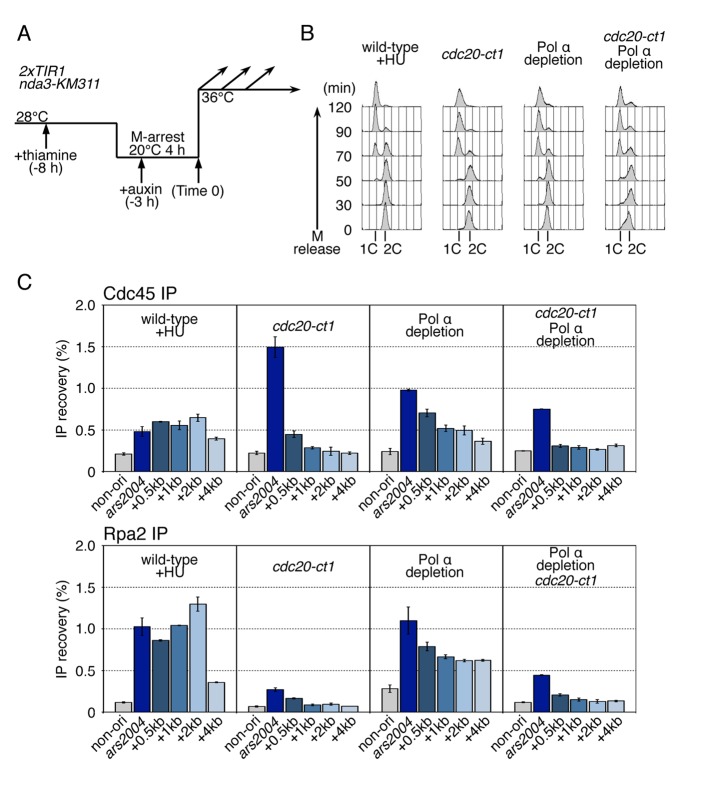
FIGURE 4:
Cdc20 CTD is required for efficient progression of CMG helicase under Pol α depletion. (A) The experimental scheme for Pol α depletion used for the experiments in this figure is shown. HM3914 flag-cdc45 2xTIR1 nda3-KM311 (wild-type), HM3910 cdc20-ct1 flag-cdc45 2xTIR1 nda3-KM311 (cdc20-ct1), HM4017 Pnmt81-pol1-aid Pnmt81-spp2-aid flag-cdc45 2xTIR1 nda3-KM311 (Pol α depletion), and HM4095 cdc20-ct1 Pnmt81-pol1-aid Pnmt81-spp2-aid flag-cdc45 2xTIR nda3-KM311 (cdc20-ct1 Pol α depletion) strains grown at 28°C were cultured in the presence of thiamine (10 μg/ml) for 4 h and arrested at M-phase by incubation at 20°C for 4 h, and then released at 36°C (Time 0). Auxin (0.5 mM) was added 3 h before release. HU (15 mM) was added upon release in the case of the wild type. (B) Aliquots taken at the indicated time points were analyzed by flow cytometry. Positions of 1C and 2C DNA contents are shown. (C) Samples taken at 50 min after M release were analyzed by ChIP assays. Chromatin-immunoprecipitated DNAs with anti-FLAG or anti-Rpa2 were analyzed by qPCR using primers for ars2004, the regions located 0.5 kb, 1 kb, 2 kb, and 4 kb from ars2004, denoted as “+0.5kb,” “+1kb,” “+2kb,” or “+4kb,” and the nonorigin region. Mean ± SD obtained from multiple measurements in qPCR is presented.