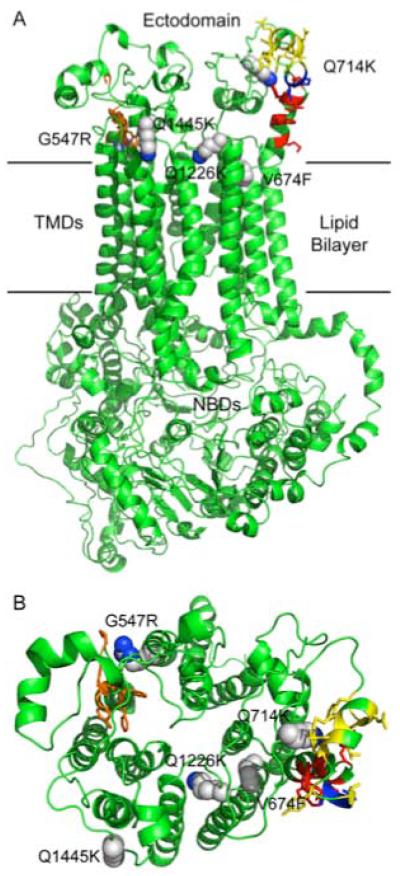
Fig. 9.
A map of RC21 suppressor mutations using a CaCdr1p homology model. The open (no ATP bound) CaCdr1p model, viewed from the side (A) and from the periplasm (B), was generated in Modeller 9v7 (Eswar et al., 2006) using the S. cerevisiae Pdr5p model of Rutledge and colleagues (Rutledge et al., 2010). A sequence alignment was generated for CaCdr1p and ScPdr5p using clustalW. Coordinates for the Pdr5p model were kindly provided by Robert Rutledge. Conserved amino acid sidechains of the PDR motif A are shown in red, the connecting sequence in blue and the PDR motif B in yellow. Conserved amino acid sidechains of the EL6 hydrophobic motif are shown in orange. Amino acid side chains of 5 of the suppressor mutants are shown as spheres, with carbons in grey and nitrogens in blue.