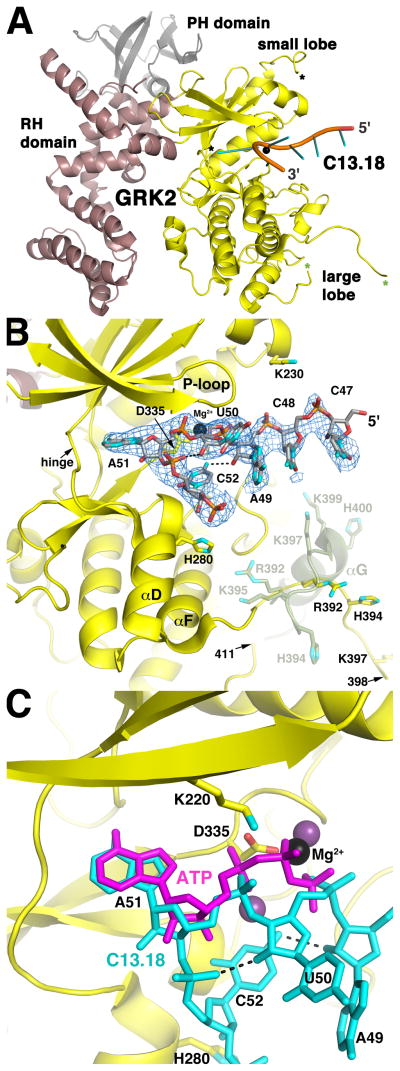
Figure 1. The C13.18 Aptamer Binds in the Active Site of GRK2.
(A) Overall structure of the C13.18-GRK2 complex. Residues 47-CCAUAC-52 of C13.18 are depicted as a cartoon ribbon with spokes indicating nucleotide bases. The kinase, RH and PH domains of GRK2 are shown in yellow, burgundy and grey, respectively. Mg2+ is a black sphere. Asterisks demark residues flanking disordered regions of the kinase domain: a segment of the active site tether (amino acids 475 and 492, black) and a segment spanning the αG helix of apo-GRK2 structures (amino acids 398 and 411, green). Aptamer binding induces an 8 change in the orientation of the large and small lobes (Figure S3A).
(B) Interactions of the C13.18 aptamer in the active site of GRK2. The blue wire cage corresponds to a 2m|Fo|−D|Fc| simulated annealing omit map contoured at 1 σ around residues 47-CCAUAC-52 of C13.18. Carbons atoms are grey for C13.18, and yellow for the kinase domain. Nitrogens are cyan, oxygens red, phosphates orange and Mg2+ black. Intramolecular hydrogen bonds in the RNA are dashed lines. The position of the αF-αG loop and αG helix in apo-GRK2 (amino acids 391–410; PDB entry 2BCJ (Tesmer et al., 2005)) is shown in green at half transparency and is reorganized in the C13.18-bound structure (see also Figure S3A). The twelve C-terminal nucleotides of the aptamer are disordered, but extend into a crystal contact lined with positively charged amino acids (Figure S3B).
(C) C13.18 mimics the binding of ATP in the GRK2 active site. GRK2-bound ATP was homology modeled based on the GRK1·2Mg2+·ATP complex (purple; PDB entry 3C4W (Singh et al., 2008)). The P-loop was omitted for clarity.