Figure 3.
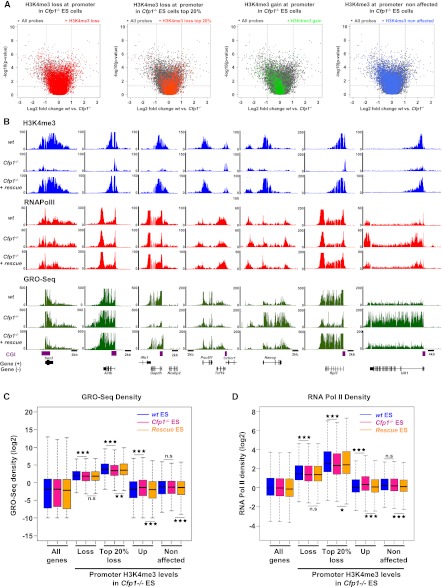
Transcription is weakly affected by decreased H3K4me3 at active gene promoters. (A) Volcano plots comparing expression values between wild-type and Cfp1−/− ES cells. Fold change in expression values (wild-type vs. Cfp1−/− ES cells) and statistical significance of the differences (two-sided t-test) were computed for each probe on the array (gray dots). Genes differentially expressed twofold or more lie above the horizontal threshold (P = 0.05) and outside the two vertical thresholds (plus or minus twofold differential expression). Colors denote genes in Cfp1−/− ES cells with decreased H3K4me3 (red), with the 20% most H3K4me3-depleted (orange), with increased H3K4me3 (green), and unaffected (blue). (B) Screenshots representing read coverage of H3K4me3 ChIP-seq signal (blue), RNA Pol II ChIP-seq signal (red), or GRO-seq signal (green) in wild-type, Cfp1−/− and wild-type rescue ES cells at selected gene loci (Sox2, Actb, Gapdh, Pou5f1, Nanog, Rpl3, and Ulk1). (C) Comparison of GRO-seq read density for wild-type (blue), Cfp1−/− (pink), and wild-type rescue (orange) at all Ensembl genes (All), genes whose H3K4me3 is decreased (Loss, n = 8527), the 20% most affected (Top 20% loss, n = 1706), genes whose H3K4me3 is increased (Gain, n = 1799), or genes whose H3K4me3 is unaffected (n = 8663) in Cfp1−/− ES cells. Box plots show the central 50% of the data (filled box), the median (central bisecting line), and 1.5× the interquartile range (whiskers). P-values were calculated using the nonpaired Wilcoxon test; (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. (D) As C, but showing the density distribution for RNA Pol II ChIP-seq reads.