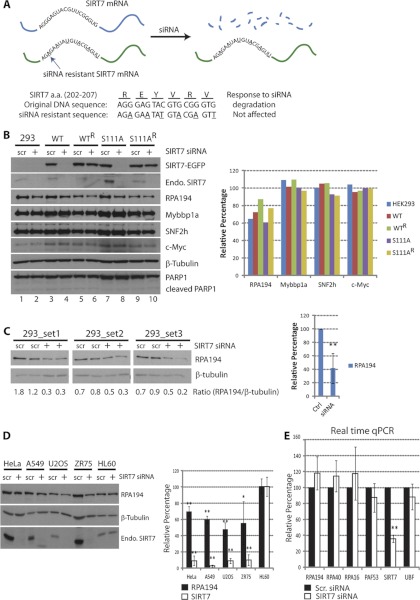
Fig. 6.
Down-regulation of RPA194 upon SIRT7 knockdown. A, diagram depicting the construction and features of the siRNA-resistant SIRT7 construct. The mRNA transcribed from resistant gene remains intact upon siRNA treatment, whereas the mRNA from the original SIRT7 sequence is degraded. Both constructs encode the same amino acids in the 202–207 region; Bottom panel, sequence difference. B, SIRT7 knockdown leads to reduced Pol I protein levels. Left panel, immunoblotting of cell lines treated with scrambled siRNA (scr) and SIRT7 siRNA (+). Right panel, relative changes in protein levels determined by densitometry and calculated as: (Iprotein/Iβ-tubulin)SIRT7 siRNA/(Iprotein/Iβ-tubulin)scrambled siRNA × 100; I = integrated intensity. C, knockdown of endogenous SIRT7 leads to reduced Pol I protein levels. Left panel, parental HEK293 cells were transfected with scrambled or SIRT7 siRNA (n = 3, two technical replicates), and Pol I protein levels were assessed by Western blot using anti-RPA194 antibodies. Right panel, averaged relative changes in Pol I levels. D, down-regulation of RPA194 upon SIRT7 knockdown is a general effect. HeLa, A549, U2OS, ZR75, and HL60 cells were transfected with scrambled (scr) or SIRT7 siRNA for knockdown of endogenous SIRT7. Immunoblotting (left panel) and quantification of changes in protein levels were performed (n = 3) as in B. E, down-regulation at the transcriptional level is not responsible for the reduction of RPA194. mRNA from cells treated with scrambled or SIRT7 siRNA was purified and quantified by real time quantitative PCR (n = 3, four technical replicates). *, p value < 0.05; **, p value < 0.005.