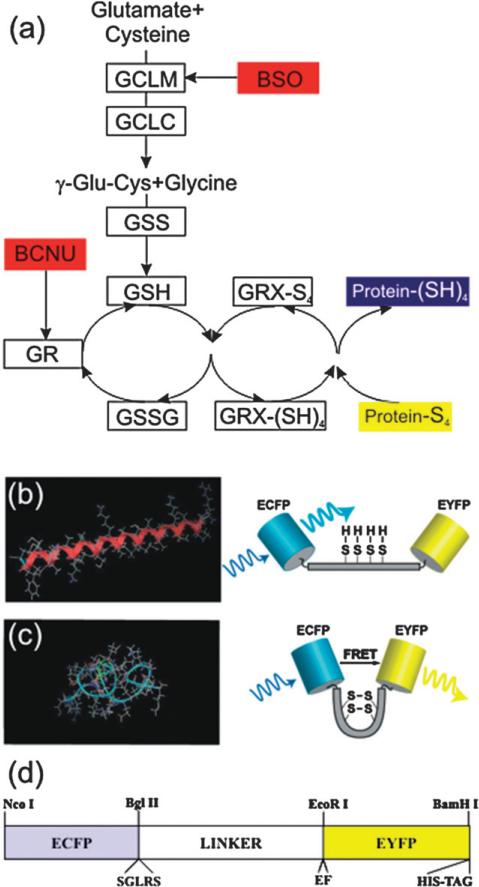
Fig. 1.
Glutaredoxin pathway of reduction of protein disulfides and a FRET biosensor to probe it. (a) The glutaredoxin system acts via the GSH/GSSG redox buffer. Its high electron-donating capacity (high negative redox potential) combined with high intracellular concentration (millimolar levels) generate great reducing power. GRX-(SH)4 and GRX-S4 indicate the reduced and oxidized forms, respectively, of glutaredoxin. In the Figure the enzymes are indicated by boxes and their inhibitors are in red. (b,c) A schematic of the redox sensitive switching mechanism for a single molecule. Under oxidative conditions, intermolecular disulfide bonds can form shifting the free energy minimum from the (b) α-helix, to a (c) ‘clamped-coil’ state (similar to a helix–coil transition). The coiled state brings the donor (ECFP) and acceptor (EYFP) in closer proximity than in the extended helix state, where they exchange excitation energy more efficiently (i.e. a high FRET state). The extent of energy transfer is easily quantified from the increased emission of the acceptor. (d) Diagram of the CY-RL7 construct, indicating the location of the redox linker inserted between two GFP variants, ECFP and EYFP. The position of the restriction sites for NcoI, BlgII and BamHI, EcoRI used for cloning are also indicated in the map along with the location of the hexa histidine-tag, identified by the trademarked name His-tag, which is used for protein purification.