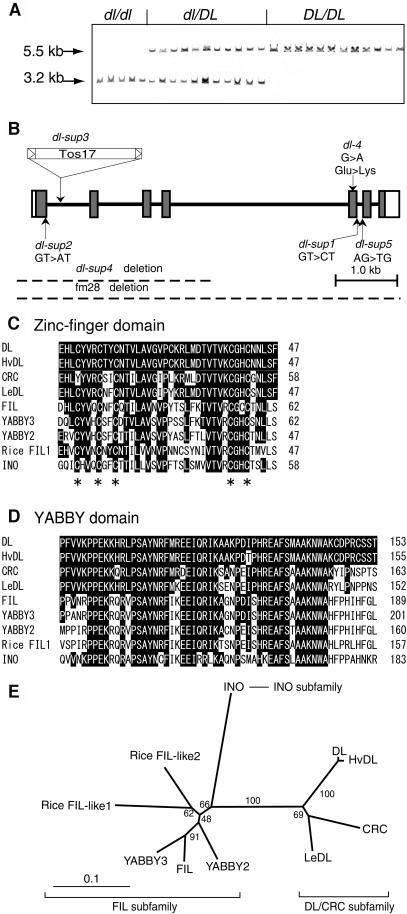
Figure 3.
Gene Isolation and Structural Features of DL.
(A) Cosegregation of the TOS17 insertion with DL genotypes. The genotypes, which were determined by the drooping leaf phenotype of the next generation (R3), are shown above the lanes. The 5.5-kb XbaI band corresponds to the wild-type allele, whereas the 3.2-kb band is produced by the insertion of TOS17, which contains an internal XbaI site.
(B) Genomic structure of DL and mutations in the six dl alleles. Boxes indicate exons and thick lines indicate introns. The coding regions are shown by shaded boxes.
(C) Zinc-finger domain. The five conserved Cys residues are indicated with asterisks.
(D) YABBY domain. The regions containing both domains have been extended slightly beyond the original definition (Bowman and Smyth, 1999) through the accumulation of more YABBY sequences. Amino acids identical to those of DL are indicated with black boxes.
(E) Phylogeny of the YABBY gene family. The tree was constructed by the neighbor-joining method (Saitou and Nei, 1987). Numbers denote bootstrap values. Amino acid sequences of HvDL (Hordeum vulgare [barley] DL homolog) and LeDL (Lycopersicon esculentum [tomato] DL homolog) were deduced from the following EST sequences: HvDL (BG369278, AL509850) and LeDL (AI485831, AI483816). Rice FIL-like proteins have been described by Sawa et al. (1999).