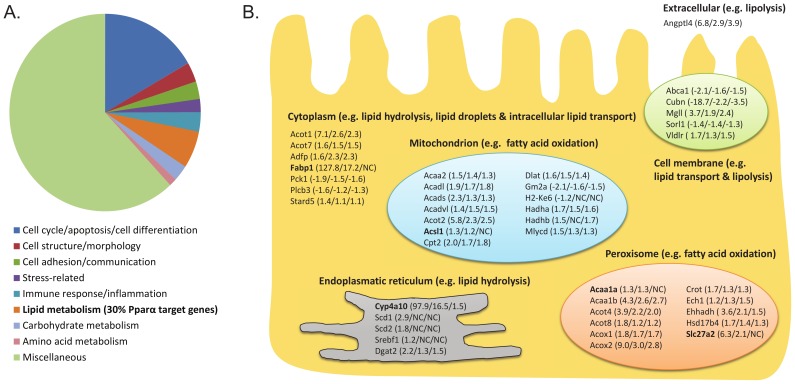
Figure 1. Effect of heme on PPARα target genes.
A. Categorization of heme-induced differentially expressed genes (q<0.01 and signal intensity>20 in at least treatment) according to GO Biological Process annotation. Figure is based on results from IJssennagger et al. [5] showing that lipid metabolism-related gene expression is substantially influenced by heme. Thirty percent of these heme-induced lipid metabolism-related genes are PPARα target genes [18]. Miscellaneous contains processes with broad and thus unspecific biological process terms. B. Expression of PPARα target genes in enterocytes is mainly upregulated. Behind the gene the fold- changes are indicated from colonic scrapings from heme fed vs. control mice from resp. the previous experiment with C57Bl6J mice, current experiment WT SV129 mice and current experiment KO SV129 mice. In bold are PPARα targets of which no significant induction is seen in the KO mice.