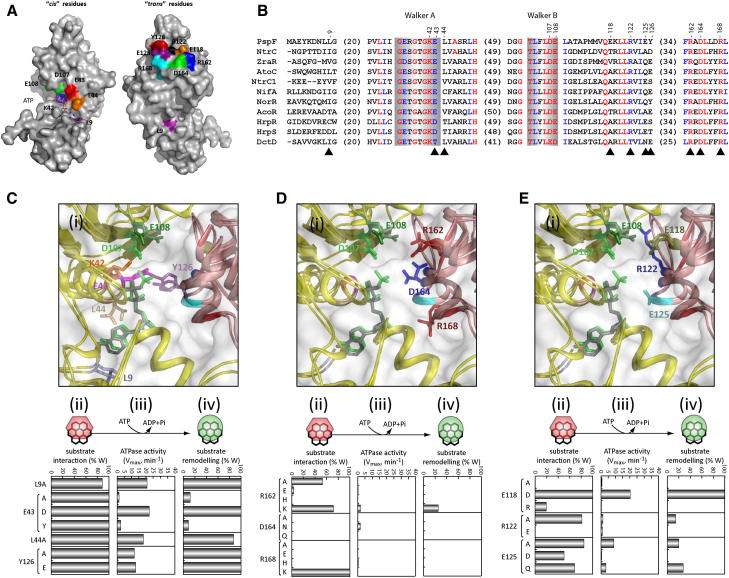
Figure 1.
Sequence Alignment, Localization, and Functional Activities of PspF Variants
(A) Localization of “cis” and “trans” residues on the PspF structure (pdb 2C9C) (Rappas et al., 2006). Figure prepared using PyMol software.
(B) Sequence alignment of PspF from Escherichia coli, NtrC from E. coli, ZraR from E. coli, AtoC from E. coli, NtrC1 from Aquifex aeolicus, NifA from Sinorhizobium meliloti, NorR from E. coli, AcoR from Pseudomonas aeruginosa, HrpR and HrpS from Pseudomonas syringae pv. tomato str. DC3000, and DctD from Rhodobacter capsulatus. Numbering is based on PspF sequence. Black triangles: amino acid substitution of the variants studied.
(C–E) Side-chain orientation in the presence of ATP or ADP (i), substrate interaction activity by quantifying the amount of stable σ54-PspF complex formed in the presence of ADP-AlF (ii), ATPase activity (iii) (see Table 1 for detail), and substrate remodeling activity by using RPo formation assay (iv) were tested for L9, E43, L44, and Y126 variants (C); R162, D164, and R168 variants (D); and E118, R122, and E125 variants (E). Experiments were performed at least in triplicate, and the maximal variation observed was lower than 10%.