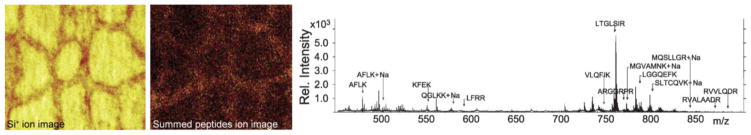
Figure 2. Static SIMS imaging of a large protein at cell-scale resolution.
Thyroglobulin (660 kDa) in thyroid gland tissue is visualized using static matrix-enhanced SIMS after on-tissue trypsin digest. An Si+ ion image (left) of sectioned tissue reveals cell morphology since removal of colloid within the cells exposes the underlying silicon substrate. Summed signals of the detected tryptic peptides generates an ion image (middle) indicating protein localization along the epithelial cell borders. A mass spectrum (right) from the tissue on the right shows labeled tryptic peptides in the m/z 450–900 range. Images are represented in false-color scale ranging from black (low signal) through red to yellow (high signal); field of view is 500 × 500 μm. Adapted with permission from ref. [87], copyright 2010, John Wiley and Sons.