Abstract
Studying the functional architecture of the brain requires technologies to precisely measure and perturb the activity of specific neural cells and circuits in live animals. Substantial progress has been made in recent years to develop and apply such tools. In particular, technologies that provide precise control of activity in genetically defined populations of neurons have enabled the study of causal relationships between and among neural circuit elements and behavioral outputs. Here, we review an important subset of such technologies, in which neurons are genetically engineered to respond to specific chemical ligands that have no interfering pharmacological effect in the central nervous system. A rapidly expanding set of these “orthogonal pharmacogenetic” tools provides a unique combination of genetic specificity, functional diversity, spatiotemporal precision, and potential for multiplexing. We review the main classes of orthogonal pharmacogenetic technologies, including neuroreceptors to control neuronal excitability, systems to control gene transcription and translation, and general constructs to control protein–protein interactions, enzymatic function, and protein stability. We describe the key performance characteristics informing the use of these technologies in the brain, and potential directions for improvement and expansion of the orthogonal pharmacogenetics toolkit to enable more sophisticated systems neuroscience.
Keywords: Pharmacology, pharmacogenetics, neural control, systems neuroscience, brain interfaces, optogenetics
Introduction
The brain is a complex system comprising billions of interconnected, specialized cells whose collective function gives rise to mental states and observable behavior, while malfunction leads to neurological and psychiatric disease. Studying this system requires technologies to precisely sense and control the activity of specific neural cells and circuits in model organisms. An important focus of technical development in recent years has been technologies that provide precise control of activity in genetically defined populations of neurons. Such technologies have enabled the study of causal relationships between the functioning of neural circuits and behavior, yielding novel insights into processes such as aggression,1 anxiety,2 and appetite.3 Here, we review an important subset of such technologies, in which exogenous genes introduced into neurons enable them to respond to specific chemical ligands that have no interfering pharmacological effect in the central nervous system (CNS). An expanding repertoire of such tools provides a powerful combination of genetic specificity, functional diversity, spatiotemporal precision, and potential for multiplexing that will be critical in obtaining a systems-level understanding of brain function.
In the past, neuroscientists have modulated neural activity using pharmacology or electrical stimulation, obtaining either molecular or spatial specificity (Table 1). Each method is incomplete, since both location and molecular identity are needed to define the functional circuit roles of neurons. Recently, novel technologies have been developed that are capable of controlling neural activity with both spatial and molecular precision. These technologies take advantage of advances in understanding of cell-type-specific gene expression in neurons4 and methods of targeting transgenes to cells based on their genetic properties, location, and circuit connectivity.5 Control is achieved by expressing exogenous actuator proteins that make specific neurons responsive to “orthogonal” stimuli that normally have no effect on nervous system function.
Table 1. Capabilities of Neural Control Technologiesa.
| conventional pharmacology | electrical stimulation | optogenetics | orthogonal pharmacogenetics | |
|---|---|---|---|---|
| cell type specificity | medium | none | high | high |
| temporal precision | medium | high | high | medium |
| spatial precision | low | high | high | medium |
| multiplexing | low | low | medium | high |
| signaling variety | low | low | medium | high |
| spatial coverage | high | low | low | high |
| requires gene delivery | no | no | yes | yes |
| requires device | no | yes | yes | no |
Positive characteristics in bold. Negative characteristics in italic.
One successful expression of this concept, “optogenetics”, uses actuator proteins that are sensitive to visible light, including ion channels, transporters, G-protein coupled receptors (GPCRs), and protein–protein binding domains. Expressing these proteins in neurons makes it possible to control various aspects of their activity with light.6 In addition to the molecular, spatial, and circuit specificity achievable through genetic targeting, optical stimulation provides a high degree of temporal precision, in some cases on millisecond time scales, enabling control of neuronal spike timing and frequency7 (Table 1). Multiplexing is possible with up to three or four channels using actuator proteins that respond to different wavelengths. A drawback of optogenetic brain stimulation in mammals is the need for implanted optical fibers to deliver light. In addition to being burdensome experimentally, the resulting localized illumination makes it difficult to control diffuse signaling networks.
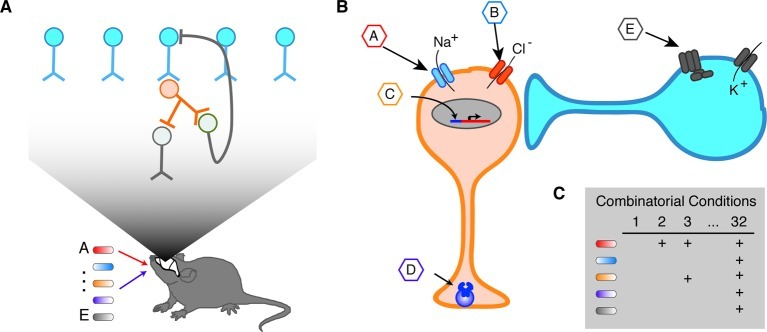
Another approach to orthogonal control of genetically specified neurons uses actuator proteins that respond to unique chemical ligands that have no interfering pharmacological activity in the CNS. We refer to this approach as “orthogonal pharmacogenetics” (OP). While “pharmacogenetics” on its own has been used by other workers in this field, we believe “orthogonal” helps distinguish this class of technology from clinical pharmacogenetics, the study of endogenous genetic polymorphisms affecting individual response to drugs. OP has been used for some time to control gene expression (e.g., using tetracycline-dependent transcriptional promoters). Recently, novel actuator proteins have been developed that enable chemical control of neuronal firing, second-messenger signaling, and synaptic function. Like optogenetics, OP relies on genetic targeting to achieve molecular, spatial, and circuit specificity. In addition, ligands with different pharmacokinetic properties can be used to specify the time scale of neural control, ranging from minutes to days. This temporal resolution is not as high as that of optogenetics. However, it is fully satisfactory in many cases where circuits play modulatory roles or the objective of the perturbation is long-term inhibition. Unlike optogenetics, OP does not require invasive implants, and both local and diffuse groups of neurons can be controlled depending on where the actuator gene is expressed (Table 1). In theory, OP also has the capacity for virtually unlimited multiplexing, as long as a sufficient number of unique ligand–receptor pairs can be developed. Importantly, such multiplexing can be both within a cell type (e.g., by expressing inhibitory and excitatory ion channels controlled by different ligands) and between multiple cell types (Figure 1).
Figure 1.
Illustrated example of multiplexed orthogonal pharmacogenetics. (A) Two cell types (blue and orange) involved in a particular neural circuit (top) are genetically modified to express orthogonal actuators responding to several distinct ligands that can be administered orally to the model organism (bottom). (B) One neuron (orange) expresses four distinct OP constructs, enabling temporally specific, multiplexed control of excitation (ion channel controlled by ligand A), inhibition (ion channel controlled by ligand B), gene transcription (transcriptional transactivator controlled by ligand C), and decreased presynaptic transmitter release (vesicle protein multimerization controlled by ligand D). A second neuron (blue) has an orthogonal GPCR coupled to an endogenous potassium channel, enabling orthogonal inhibition under control of ligand E. (C) Using the five ligands corresponding to different orthogonal actuators, it is possible to test 32 binary (ligand on or off) experimental conditions in this system.
OP systems have been engineered to provide chemical control over various aspects of neural activity, including ion channel and GPCR signaling, gene transcription, and synaptic function. In addition, OP actuators have been developed that provide control over gene translation and enzymatic activity that could be adapted to neurons. Below, we highlight the major categories of recently developed OP systems and their applications in neuroscience. We evaluate them with reference to a common set of performance characteristics applicable to functional actuators (orthogonality, compatibility, modularity, and deliverability), their chemical effector ligands (molecular specificity and deliverability), and the combination of ligand and actuator (temporal response, dose response), as defined in Table 2.
Table 2. Performance Characteristics of Orthogonal Pharmacogenetic Systems.
| Actuator Characteristics | |
| orthogonality | actuator is insensitive to endogenous ligands or other signaling elements; actuator is inactive until triggered by ligand (or inactive in presence of ligand in a switch-off system) |
| compatibility | endogenous machinery needed for actuator performance is present in target cells; actuator does not interfere with normal cell function unless it is activated by ligand |
| modularity | actuator can be modified to produce different signaling effects upon ligand binding |
| deliverability | actuator can be delivered to target cells by viral vectors and through transgenesis; ideally, the essential genetic payload should be a single gene smaller than ∼4.5kb to enable single AAV construct delivery |
| Effector Ligand Characteristics | |
| molecular specificity | at the effective dose, ligand acts only on its corresponding actuator |
| deliverability | ligand is bioavailable, preferably per orum, and penetrates CNS |
| System Characteristics | |
| temporal response | on and off kinetics for cellular and behavioral response after administration as determined by ligand pharmacokinetics and receptor activation, inactivation, and second-messenger signaling |
| dose response | dependence of cellular and behavioral response on ligand dose |
Orthogonal Neuroreceptors: Ion Channels and GPCRs
The most active recent area of development in OP has focused on neuroreceptors. Both ligand-gated ion channels (LGICs) and GPCRs have been developed as orthogonal actuators by identifying or engineering receptors with minimal sensitivity to endogenous neurotransmitter agonists and strong activation by specific exogenous ligands that have no other significant pharmacological effect in the CNS. Targeted expression of these orthogonal receptors permits temporally controlled excitation or inhibition of neurons through the administration of their cognate ligands.
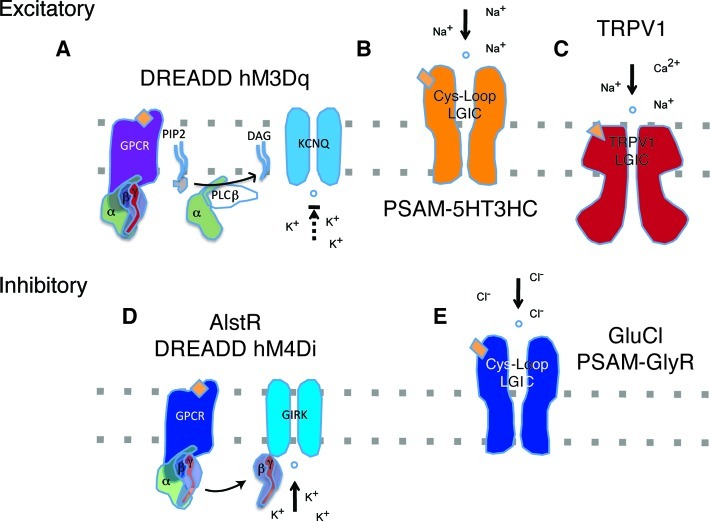
The first orthogonal GPCR and LGIC systems for use in neuroscience were based on receptors from nonmammalian organisms. The Callaway group developed a system based on the Drosophila allatostatin receptor (AlstR) and its cognate neuropeptide ligand allatostatin (AL), neither of which is expressed in mammals.8 AL does not cross-activate endogenous mammalian GPCRs, nor is AlstR activated by mammalian GPCR ligands.9 Activation of heterologously expressed AlstR by AL leads to Gi-coupled activation of endogenous mammalian G protein-gated inward rectifier K+ (GIRK) channels, leading to a reduction in cell excitability (Figure 2). Virally targeted expression of AlstR in cortical and thalamic neurons and intracranial administration of AL produce neuronal silencing on a time scale of minutes in several species.10
Figure 2.
Mechanisms of orthogonal neuroreceptors. GPCRs form the basis for both excitatory and inhibitory OP systems (A, D) based on interactions with different endogenous G proteins. GPCR signaling cascades leading to excitation and inhibition are described in the text. Cys-loop LGICs (B, E) are also used to effect inhibition and excitation based on pore domain ion selectivity. TRPV1 (C) excites cells through a nonselective cation conductance.
Around the same time, the Lester group adapted the Caenorhabditis elegans glutamate-gated chloride channel (GluCl) for silencing of mammalian neurons by administration of the anthelmintic GluCl agonist ivermectin (IVM). GluCl was rendered insensitive to its native ligand glutamate by a single point mutation and codon-optimized to achieve greater expression in mammalian cells.11 IVM activation of GluCl α and β subunits expressed in neurons elicits a Cl– conductance across the membrane that effectively shunts action potential generation (Figure 2). The GluCl/IVM system later became the first to be used for neuronal silencing with a systemically administered ligand in awake, behaving animals.12
More recently, versatile orthogonal neuroreceptor systems have been established by modifying mammalian GPCRs and LGICs. A collection of modified GPCRs called DREADDs, “designer receptors exclusively activated by designer drugs”, were developed using a combination of directed evolution and rational protein engineering.13 Building on previous efforts to engineer the ligand selectivity of GPCRs,14 the first DREADDs were generated from the human M3 muscarinic receptors (hM3). Survival screens based on the yeast pheromone response15 were used to evolve this receptor for activation by the small molecule clozapine-N-oxide (CNO) and lack of activation by the native ligand acetylcholine. CNO is a normally inactive metabolite of the atypical antipsychotic clozapine. CNO activation of the mutant hM3D triggers Gq-coupled signaling leading to membrane depolarization through phospholipase Cβ (PLCβ)/PIP2 mediated inhibition of KCNQ channels16 (Figure 2). Following a similar design scheme, a second CNO-activated DREADD, hM4D, was generated that couples to Gi, leading to activation of GIRK channels and neuronal silencing similar to that elicited by AlstR/AL (Figure 2).
Recently, a systematic engineering approach was also taken to the development of a modular system of orthogonally controlled Cys-loop ion channels with distinct ligand sensitivity and ion conductance properties.3a The modularity of this system is based on fusing the α7 nicotinic acetylcholine receptor (nAChR) ligand-binding domain onto the ion pore domain of either a cation-selective serotonin 5-HT3 receptor (α7–5HT3) or anion-selective glycine receptor (α7–GlyR) to produce functional channels with the same pharmacological profile but different ion permeability.17 Novel ligand recognition properties were engineered through a “bump-hole” approach, which uses structural models to generate libraries of predicted ligand–receptor pairs that are then synthesized and screened for selective functional activity. Structural analogs of the α7-specific synthetic agonist PNU-282987 were tested for selective activation of mutant, but not wild-type, channels. At the same time, mutant channels were screened for lack of activation by acetylcholine and nicotine. The resulting mutant ligand binding domains are dubbed “pharmacologically selective actuator modules” (PSAMs). Each PSAM is exclusively activated by a cognate synthetic agonist, called a “pharmacologically selective effector molecule” (PSEM). Three specific PSAM/PSEM tools have been designed, each with different ion conductance properties for controlling neuronal excitability.3a These include the cation-selective activator, PSAMQ79G,Q139G–5HT3HC/PSEM22S, the anion-selective silencer, PSAML141F,Y115F–GlyR/PSEM89S, and a third Ca2+-selective channel, PSAMQ79G,L141S–nAChR V13′T/PSEM9S.
Another orthogonal LGIC system is based on the transient receptor potential ion channel TRPV1, an endogenous mammalian receptor predominantly expressed in the peripheral nervous system. TRPV1 is a nonselective cation channel activated by noxious heat, pH, and exogenous ligands including the hot chili pepper compound capsaicin.18 Targeted neuronal expression of TRPV1 in the mouse brain leads to capsaicin-activated currents and action potentials.19 To use TRPV1 for orthogonal control of specific neurons, the host organism can be modified to knock out endogenous TRPV1 expression. On this TRPV1–/– background, one can reintroduce TRPV1 into target cells as an exogenous OP actuator.20 This approach also works in model organisms that naturally lack TRPV1 receptors.21 Background modification has also been used to create orthogonal inhibition using the GABAA receptor. A single point mutation in its γ2 subunit makes this Cl–-permeable LGIC insensitive to the synthetic agonist zolpidem. In homozygous mice carrying this mutation, targeted restoration of the wild-type subunit makes a select group of neurons zolpidem-responsive.22
Performance Characteristics
The set of available OP neuroreceptor tools is summarized in Table 3. Their specific performance characteristics inform their ability to fulfill the unique objectives of a neuroscience study. As defined in Table 2, key performance characteristics depend on the properties of actuators, effectors, or both.
Table 3. Orthogonal Neuroreceptors.
| class | actuator | effector | effect on neurons | signaling and endogenous partners | design and proof-of-concept | applications |
|---|---|---|---|---|---|---|
| GPCR | AlstR (Drosophila) | AL | decreased excitability | Gi-coupled; activates GIRK K+ channel | (8,10,32) | (38,39) |
| GPCR | DREADD hM4Di (human) | CNO | decreased excitability | Gi-coupled; activates GIRK K+ channel | (13) | (3b,40,41,43) |
| GPCR | DREADD hM3Dq (human) | CNO | increased excitability | Gq-coupled; inhibits KCNQ K+ channel | (13,16) | (3b,42,43) |
| LGIC | GluCl (C. elegans) α and β subunits | IVM | usually inhibition | Cl– channel | (11,12,80) | (1,27) |
| LGIC | PSAM–5HT3HC (human–mouse) | PSEM22S | excitation | cation channel (Na+ ≈ K+ > Ca2+) | (3a) | |
| LGIC | PSAM–GlyR (human) | PSEM89S | usually inhibition | Cl– channel | (3a) | (3a) |
| LGIC | PSAM–nAChR V13′T (human–rat) | PSEM9S | not shown | Ca2+ channel | (3a) | |
| LGIC | TRPV1 (rat) | capsaicin | excitation | cation channel (Ca2+ > Na+ ≈ K+) | (19−21) | |
| LGIC | GABAA (mouse) | zolpidem | usually inhibition | Cl– channel | (22) |
Actuator Orthogonality, Compatibility, Modularity, and Deliverability
GPCR and LGIC architectures of orthogonal receptors confer distinct functional properties. Neural control using GPCR-based systems depends on second messenger signaling cascades. Although these secondary effectors are generally present in neurons, their precise quantity and subcellular localization could impose limits on actuator function. Conversely, expression of heterologous receptors could sequester second messenger molecules, disrupting endogenous receptor activity.23 G-protein-mediated cascades may also have undesirable effects beyond altering neuronal firing (e.g., affecting gene expression), especially with sustained activation.24 In contrast to GPCRs, LGIC actuators are self-contained membrane proteins with ligand-dependent ionic conduction directly affecting membrane excitability. They require no intermediary molecules. However, close attention must be paid to their ionic selectivity. The high Ca2+ permeability of TRPV1, for example, is likely to trigger Ca2+-mediated cell signaling events in addition to exciting cells.
Both LGICs and GPCRs are functionally modular. The PSAM/PSEM system described above illustrates the relative ease of generating new chimeric channels based on the modularity of Cys-loop receptors. Ligand-binding domains developed and tested while connected to one transmembrane domain were transplanted onto other transmembrane domains, resulting in constructs with completely different ionic conductance. Structure–function studies support further potential for altering ion selectivity, single-channel conductance, and open channel duration (reviewed in ref (25)). When modifying receptors, one must ensure that mutant channels have minimal leak current in the resting state. GPCRs are modular with regard to their second messenger coupling. Domain swapping and point mutations of intracellular loops can alter G-protein specificity, allowing modulation of Gi-, Gs-, and Gq-coupled signaling pathways.26
Engineered receptors can be delivered into the CNS via transgenic modification or viral vectors. With coding sequences of approximately 1.7 kb for the M3 muscarinic receptor, 1.2 kb for AlstR, 1.4 kb for GluCl, 1.5 kb for PSAMs and 2.5 kb for TRPV1, each receptor construct can be accommodated by lentivirus and adeno-associated virus (AAV) vectors. Most of these tools require the delivery of only one genetic construct, except GluCl, which requires α and β subunits. The requirement for two constructs permitted GluCl to be used with intersectional genetic targeting.1,27 Codon optimization and signal peptide fusions can improve translation and membrane trafficking of non-native receptors.11b,28 Receptors can also be regionally targeted to somatodendritic, axonal, or postsynaptic sites.29
Ligand Deliverability and Specificity
Ligands with good pharmacokinetics, including oral bioavailability and brain penetration, allow manipulation of deep brain structures and dispersed neuronal populations. The ability to conveniently deliver effector ligands orally or by intraperitoneal or intravenous injection is a key advantage of the DREADD/CNO, GluCl/IVM, and the PSAM/PSEM systems (Table 4). On the other hand, neuronal manipulation using AlstR/AL or TRPV1/capsaicin (in a wild-type background) requires localized application of effector ligands via parenchymal or intracerebroventricular administration. AL is a neuropeptide that cannot cross the BBB. In wild-type background, systemically administered capsaicin would elicit unwanted effects via endogenous TRPV1 receptors.
Table 4. Orthogonal Control of Gene Expression and Intracellular Signaling.
| control target | design | actuator examples | effector examples | uses in CNS | refs |
|---|---|---|---|---|---|
| DNA transcription | ligand-dependent recombinase | CreER, CrePR, CreGR, FlpER | tamoxifen, RU486, dexamethasone | numerous; reviewed in refs (50,54) | (53,56−58) |
| DNA transcription | ligand-dependent transcription factor | tTR, rtTR, GLVP | doxycycline, RU486 | numerous; reviewed in refs (50,54) | (51,52,55) |
| RNA translation | riboswitch | self-cleaving aptamer | toyocamycin | (65,66) | |
| dimerization-activated signaling | dimerizing domains fused to target proteins | fusions with FKBP, cyclophilin, FRB, DHFR, or gyrase B | dimerizers based on FK506, cyclosporin, rapamycin, methotrexate, or coumermycin | ligand-dependent suppression of long-term potentiation using FKBPF36V–protein kinase R fusion | (67,68,70−73,79) |
| enzymatic function | insertion or fusion of ligand-binding domain with enzyme | maltose-binding protein-β-lactamase | maltose, sucrose | (47b,76) | |
| protein stability | ligand-stabilized degradation domains fused to protein | fusions with destabilized mutant FKBP or DHFR | FK506 derivatives, trimethoprim | ligand-dependent YFP fluorescence in striatum using mDHFR–YFP | (77,78) |
| synaptic signaling | dimerization inactivation of vesicle fusion proteins | fusion of VAMP2 with FKBPF36V | AP20187 (FK506 dimer derivative) | ligand-dependent inhibition of synaptic signaling in Purkinje cells | (75) |
To achieve truly orthogonal control, effector ligands must have no significant activity in cells not expressing their partner actuator at doses used for actuation. IVM is known to activate or potentiate other Cys-loop receptors present in the CNS, but with much lower sensitivity.30 PSEMs were screened for ligand binding by radioligand displacement against a number of other LGICs, GPCRs, and transporters,3a revealing weak to moderate binding of PSEM89S to the α4β2 neuronal nAChR receptor; off-target functional activation remains to be assayed. Conversely, undesired on-target effects can result from agonism by endogenous ligands. For example, endogenous TRPV1 ligands including the endocannabinoid anandamide and N-arachidonoyl-dopamine are expressed in the CNS31 and could possibly allow capsaicin-independent enhancement of neuronal activity. For each system, it is important to determine an effective dosage range for optimal control with minimal side effects.
Temporal Resolution and Dose Response
The activation and deactivation kinetics of in vivo neuronal manipulation using OP systems can range from minutes to hours and depend on the pharmacokinetic properties of the ligand such as absorption, distribution, metabolism, and excretion, as well as receptor properties including affinity for agonist, desensitization, and internalization. The TRPV1/capsaicin tool allows the most rapid transient neuronal activation, with excitatory responses occurring within minutes of administration and lasting approximately 10 min, attributed to rapid capsaicin metabolism.20 Activation of DREADDs by CNO can also be observed within 5–10 min of drug administration, with induced behavior lasting from minutes to longer than 9 h. GPCRs are especially sensitive to desensitization or internalization with prolonged ligand exposure. These processes can either terminate a pharmacologically induced signal prematurely or facilitate sustained signaling or hyperexcitability32 because endocytosis of GPCRs does not always terminate the signal.33 IVM-induced GluCl currents activate over several hours and remain open for periods on the order of 8 h, presumably because neither desensitization nor ligand dissociation occur. Silencing effects by GluCl/IVM can last for 2–4 days; postsilencing recovery may require receptor turnover.12 Long periods of enhanced or silenced activity can be beneficial in some experiments, but present the risk of adaptative, compensatory, or plastic changes at the cellular or network levels. PSAMs are activated by their ligands within 15 min of administration, and recovery is observed after 24 h. PSEM22S and PSEM89S have brain half-lives of 40–80 min following intraperitoneal injection.3a
Where temporal response depends on desensitization kinetics, it may be possible to modify it at the actuator level. Mutations in the ligand binding domain, transmembrane domains, and large cytoplasmic domain of Cys-loop receptors have all been shown to affect desensitization.34 For TRPV1, a point mutation that reduces Ca2+ permeability also abolishes desensitization.35 Phosphorylation is also known to effect desensitization of many membrane receptors.35,36 The removal of phosphorylation sites in the C-terminus of heterologously expressed GPCR produced receptors that were resistant to internalization and less prone to desensitization, resulting in prolonged signaling.37 For applications requiring more defined end points, it may be possible to design synthetic antagonists or selective pore blockers for controlled termination of manipulated activity. Thus, there would be both an “on” ligand and an “off” ligand.
Dose-dependence of behavioral responses has been reported for Alst/AL38 and GluCl/IVM,12 and dose-dependent increases in neuronal activity have been demonstrated with hM3Dq/CNO16 and TRPV1/capsaicin.19,20 There is no in vivo dose–response information for the PSAM/PSEMs.
Applications of Orthogonal Neuroreceptors
Several orthogonal neuroreceptor systems have been used in vivo to study neural circuitry, and we provide examples of their use in mammals (Table 3). Viral-mediated expression of AlstR has been targeted to somatostatin-expressing neurons of the ventrolateral medulla to study pathological breathing patterns of adult rats.38 Transgenic mouse lines expressing AlstR have been used to examine locomotor activity in V1 and V3 spinal cord neurons.39
GluCl/IVM-induced silencing has been used in conjunction with channelrhodopsin-2 (ChR2) mediated activation to define an inhibitory microcircuit within the amygdala involved in mouse fear conditioning.27 Because the GluCl channel requires coexpression of α and β subunits, an intersectional approach was used to restrict the expression of GluCl to specific GABAergic neurons within an anatomically defined amygdala subregion. The GluCl/IVM system has also been used to study a hypothalamic locus involved in male mouse aggression and mating behaviors.1
Viral vectors bearing different gene promoters have been used for targeted expression of the hM4Di/CNO DREADD silencer in striatonigral vs striatopallidal neurons to study the opposing roles of direct and indirect pathways in regulating adaptations from repeated psychostimulant drug exposure.40 This system was also used to study the role of serotonergic neurons in respiration and thermoregulation.41 Recently, the hM3Dq/CNO activator was expressed in an activity-dependent manner to examine how artificial reactivation of a stimulated network affects the encoding of contextual fear memory in mice.42 The hM4Di/CNO silencer and hM3Dq/CNO activator tools have also been used in parallel experiments to study the opposing impact of activation and silencing of agouti-related protein (AgRP) neurons of the hypothalamus on feeding patterns and energy expenditure.3b Controlled activation and inhibition of orexinergic neurons in the hypothalamus elucidated their role in controlling sleep and wakefulness.43 Because CNO activates both excitatory and inhibitory DREADD actuators, opposite effects had to be studied in separate cohorts of animals.
Simultaneous bidirectional control of neuronal activity has been demonstrated by OP and optogenetic actuators in the same set of cells. A bicistronic Cre-dependent AAV was used to coexpress the PSAML141F,Y115F–GlyR silencer and the light-activated channel ChR2 in AgRP neurons. Voracious feeding behavior evoked from continuous photostimulation was strongly suppressed by intraperitoneal administration of PSEM89S.3a Such bidirectional modulation will be most informative for deciphering neuronal networks and their role in behavior.
Prospects for Further Engineering of Orthogonal Neuroreceptors
The systems described above represent a promising start for the use of OP to control neural activity, demonstrating actuation of various aspects of neuronal signaling over a range of time scales, triggered conveniently by peripheral ligand administration. Substantial further work is needed to enact the vision presented in Figure 1. Multiplexed control over a significant number of cell types will require a larger set of orthogonal ligand–receptor pairs. Investigators should be able to choose among OP systems with various temporal profiles to meet experimental requirements. More precise control over cellular signaling also necessitates greater “cassette” modularity of ligand interaction and signaling domains.
Further development of OP neuroreceptor systems will be aided by increasing knowledge about receptor structure. The three-dimensional structures of a number of GPCRs and Cys-loop receptors have now been resolved, including the M3 muscarinic receptor44 and the GluCl channel.45 Structures have also been solved for various conformational states, mutant forms and ligand complexes.46 Growing availability of structural data along with homology modeling and docking programs will be useful in optimizing current tools and in rational construction of new ones. Already the PSAM/PSEM system has demonstrated the utility of homology-based structural information.
A major goal of future OP receptor engineering efforts should be to expand the repertoire of ligand–receptor pairs. Most ligands used to date are either active on the native receptor or close relatives of known agonists (Table 4). Many molecules with desirable properties (lack of activity on endogenous targets, high CNS penetration, rapid PK) exist outside of this constrained chemical space. Antimicrobial medications and inactive drug metabolites, for example, are sizable categories of compounds with characterized pharmacokinetics and lack of activity in mammals. An even larger repository of potential effector ligands may be found among inactive analogs of drug candidates synthesized and characterized by pharmaceutical firms during lead compound optimization.
Engineering receptors that respond to effectors dissimilar from their native ligands could build on previous accomplishments using directed evolution13 and structure-guided modification.3a Directed evolution, in particular, has been successful in altering the chemical substrate and ligand specificity of enzymes and allosteric switches.47 Directed evolution requires efficient high-throughput screens, which are available for both GPCR signaling13 and ion channel conductance.48 Furthermore, directed evolution libraries based on structure-guided recombination between homologous proteins (or domains) have been shown to enhance evolution efficiency.49 The substantial homology of receptors and ligand-binding domains within and among organisms could enable the use of homologous recombination in OP receptor engineering.
Orthogonal Control of Transgene Expression
Orthogonal pharmacogenetic control of transgene expression has been used extensively to study genes and cell types involved in various aspects of neural development and function.50 A widely used transcriptional transactivation system is based on a fusion of the Escherichia coli tetracycline repressor protein with the VP16 transactivation domain of the herpes simplex virus.51 Tetracycline-dependent activity of this protein drives the transcription of genes placed under control of the tet operator coupled to CMV promoter elements. In the original tetracycline-regulated transactivator (tTA) system, transgene expression is turned off by tetracycline or its analogs such as doxycycline (dox). In the modified reverse tTA (rtTA) system ligand administration turns transgene expression on.52
Gene expression can also be activated or inactivated permanently after ligand administration using ligand-dependent site-specific DNA recombinases. The most established system for such control is based on the fusion protein, CreER, of the bacteriophage Cre recombinase and a mutated version of the ligand-binding domain of the estrogen receptor (ER).53 In the absence of ligand, CreER is sequestered in the cytoplasmic Hsp90 complex. Upon binding of the synthetic ER ligand tamoxifen, CreER is liberated from Hsp90 and translocates to the nucleus where it effects recombination between two loxP sites. If a target gene is preceded by an in-frame stop cassette flanked by loxP sites (“floxed”), CreER activation will remove that stop cassette and enable expression of the gene. Alternatively, the gene itself can be floxed so that CreER activity leads to its deletion; more complex genetic manipulations using Cre-based systems have also been developed.54
Other systems similar to tTA/rtTA and CreER have been engineered using different functional and ligand binding domains, including GLVP (based on the transcriptional activator Gal4) for ligand-dependent transcription55 and Flp-ER (based on the Flp recombinase) for ligand-dependent recombination,56 both of which use the ER ligand-binding domain and corresponding effector ligands. In addition, recombinase systems have been generated using ligand-binding domains from the progesterone receptor (PR) (57) and the glucocorticoid receptor (GR),58 each of which responds to unique synthetic ligands. Simultaneous use of ER-, PR-, and GR-based systems may enable multiplexed control.
The most important performance characteristics of pharmacologically controlled gene expression systems are their switching behavior and dose response. Ideally, genes under pharmacological control should have zero expression in the “off” condition and high ligand dose-dependent expression in the “on” condition. The original rtTA system has a switching response (on/off) of around 200:1 in stably transfected cells59 and requires relatively high levels of dox to achieve transactivation in the brain. These characteristics have been improved by optimizing the inducible promoter to achieve a 10 000:1 switching response59 and modifying the transactivator protein (e.g., through directed evolution) to improve ligand sensitivity by a factor of 100.60 The CreER system also has very effective switching performance, although it is species-, strain-, and cell-type-dependent.61
Gene expression actuators are usually controlled by dox or tamoxifen, as discussed above. Dox is a particularly attractive effector ligand. As an antibiotic compound, it has minimal on-target effects on mammalian tissues. Dox has >80% bioavailability after oral administration and reasonable (>10% relative to serum) penetration into the CNS.62 It reaches maximal serum levels within 3 h of oral administration and has a serum half-life of 14–24 h.63 Expression of the regulated protein is detectable within 1 h of ligand administration and lasts 24–48 h.64 Tamoxifen acts on CreER through its active metabolite 4-hydroxytamoxifen to which it is converted 6–12 h after administration; recombination occurs over the following 24 h.61c
In addition to pharmacologically controlled transcription and recombination, novel methods have been developed for pharmacological control of translation in eukaryotes (reviewed in ref (65)) that may be applied in the brain. For example, a ligand-dependent ribozyme mediating RNA self-cleavage can be incorporated immediately upstream of the start codon of a gene of interest.66 In the absence of ligand, self-cleavage leads to transcript degradation and lack of gene expression. Administering the ligand (toyocamycin) abrogates RNA degradation, permitting gene translation with an induction ratio of 191 in vivo.
Orthogonal Control of Intracellular Signaling and Synaptic Function
Other important aspects of neuronal function that may be manipulated through OP are intracellular and synaptic signaling. A number of general approaches to controlling protein function and localization have been developed that may be used for this purpose, although so far there have been few examples of their use in neuroscience.
One general approach relies on chemically induced protein–protein binding. As initially demonstrated using FKBP and the homobifunctional small-molecule ligand FK1012,67 fusing a protein of interest to a ligand-inducible dimerization domain allows that protein to be dimerized on demand. Using mutants of FKBP and derivatives of FK1012 with reduced off-target effects,68 dimerization-based systems have been constructed to actuate cell-surface receptor clustering, gene transcription, and enzymatic activity (reviewed in ref (69)). Additional ligand–protein combinations have also been developed, such as cyclosporin–cyclophilin,70 rapamycin–FRB,71 methotrexate–dihydrofolate reductase (DHFR),72 and coumermycin–gyrase B subunit.73 Fusion constructs where two such binding domains are used on separate proteins can be used with heterobifunctional dimerizing ligands to enable specific heterodimerization (reviewed in ref (69)). Constructs and ligands for various forms of dimerization are now commercially available, for example, ref (74).
The first use of chemically induced dimerization in neuroscience was by Karpova et al.,75 who fused FKBP with the synaptic vesicle protein VAMP2, enabling its intravesicular cross-linking by a homobifunctional ligand to “stall” vesicle release. Ligand application effectively inhibited excitatory and inhibitory neurotransmission and silenced spontaneous neuronal activity in brain slices. Mice expressing the construct in Purkinje neurons showed impaired motor learning and task performance after intracerebroventricular (ICV) ligand administration.
Another general approach to OP control of intracellular signaling relies on allosteric modulation of protein function or stability. Fusions of ligand-binding domains with enzymes, for example, have resulted in chemically controlled catalysis.47b,76 To date, protein engineers have mostly used model ligand-binding domains and enzymes such as the maltose-binding protein, luciferase and β-lactamase for proofs of concept. However, similar approaches may be extended to control of enzymes relevant in neuronal function. Allosteric control can also be exerted over protein stability. For example, a mutant version of FKBP that is unstable and rapidly degraded in mammalian cells can be fused to a target protein, leading to rapid post-translational degradation of that protein.77 Administration of a small-molecule ligand that binds to FKBP stabilizes the construct and prevents degradation, thereby turning “on” the target protein’s activity. A recent modification of this approach using mutated DHFR and the ligand trimethoprim (TMP) was tested in the CNS,78 demonstrating stable YFP fluorescence in striatal neurons virally transfected with DHFR–YFP only after oral administration of TMP.
The in vivo performance of the OP systems actuating protein interactions, function, and stability has not been extensively evaluated. A major limitation for their use in neuroscience appears to be CNS penetration of the currently available ligands. For example, in CNS studies, the popular FKBP dimerizer AP20187 has been administered by ICV,75,79 implying poor brain distribution. Further use of these systems in neuroscience is expected to motivate greater use of ligands such as TMP that do enter the brain. Systems for orthogonal pharmacogenetic control of intracellular signaling and gene expression that may be useful in neuroscience are summarized in Table 4.
Table 5. Key Effector Ligands Used in Orthogonal Pharmacogenetic Systems.
| ligand | origins | specificity | CNS penetration | bioavailability | kinetics | refs |
|---|---|---|---|---|---|---|
| clozapine-N-oxide (CNO) | inactive metabolite of clozapine | no known activity at effective dose | yes | oral | on = 5–10 min; clearance = 2 h | (16) |
| allatostatin (AL) | natural neuropeptide | no known activity in mammals | no | injection only | on = 1–3 min; clearance = 40–60 mina (ICV) | (10,32,38) |
| ivermectin (IVM) | anthelmintic | specific up to 10× effective dose | yes | oral | on = 4–12 h;a clearance = 2–4 daysa | (12,30) |
| PSEM89s | synthetic derivative of nAChR agonist PNU-282987 | minimal binding to endogenous nAChRs | yes | oral | on = 15 min; clearance = 1–2 h | (3a) |
| capsaicin | pepper ingredient, natural TRPV1 agonist | acts on native TRPV1 receptors unless they are knocked out | yes | oral | on = 2–5 min; clearance = <15 min | (20,31) |
| doxycycline | antibiotic | minimal effects in mammals | yes | oral | on = 3 h; clearance = 14–24 h | (62−64) |
| tamoxifen | synthetic ER antagonist | minimal effects with acute administration | yes | oral | on = 6–12 h; clearance = 24–48 h | (61c,81) |
| AP20187 | derivative of FK1012 (dimer of FK506) | 1000× specificity for mutant form of FKBP | unknown | injection only (to date) | on = 20–30 min; clearance = 1–24 ha (ICV) |
Inferred from behavioral or signaling response.
Conclusions
Systems neuroscience research is now more tractable than ever thanks in part to molecular technologies enabling precise sensing and control of neural activity. We have reviewed an important class of such technologies, which provides a chemically addressable orthogonal dimension for neural control, and development of which is a highly active area of research. While a number of orthogonal pharmacogenetic tools have been used in neuroscience to great effect, many more (including those originally developed for use outside the brain) are ready for application. Future engineering efforts are expected to increase the variety of neuronal signaling pathways that can be manipulated. In addition, we believe it is particularly important to expand the repertoire of CNS-compatible ligands used in OP to enable multiplexed interrogation within and across cell types. Here, we have focused on the use of OP tools in neurons, but other relevant cell types in the brain such as glia and endothelial cells may also be targets for application.
A key feature of this class of technologies is the ability of many OP tools to be triggered noninvasively through peripheral ligand administration. The use of these tools together with new technologies for high-resolution noninvasive molecular imaging will make it possible to create complete noninvasive neural input/output systems to study brain-wide neural circuits, complementing more localized research using optical techniques. Furthermore, as gene and cell therapy make progress toward clinical acceptance, it may be possible for genetically encoded OP and noninvasive imaging technologies to help diagnose and treat neurological disease. Thus, orthogonal approaches for interfacing with the brain point in an exciting direction for both basic and clinical neuroscience.
The authors declare no competing financial interest.
References
- Lin D.; Boyle M. P.; Dollar P.; Lee H.; Lein E. S.; Perona P.; Anderson D. J. (2011) Functional identification of an aggression locus in the mouse hypothalamus. Nature 470(7333), 221–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye K. M.; Prakash R.; Kim S. Y.; Fenno L. E.; Grosenick L.; Zarabi H.; Thompson K. R.; Gradinaru V.; Ramakrishnan C.; Deisseroth K. (2011) Amygdala circuitry mediating reversible and bidirectional control of anxiety. Nature 471(7338), 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Magnus C. J.; Lee P. H.; Atasoy D.; Su H. H.; Looger L. L.; Sternson S. M. (2011) Chemical and genetic engineering of selective ion channel-ligand interactions. Science 333(6047), 1292–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Krashes M. J.; Koda S.; Ye C.; Rogan S. C.; Adams A. C.; Cusher D. S.; Maratos-Flier E.; Roth B. L.; Lowell B. B. (2011) Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J. Clin. Invest. 121(4), 1424–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatten M. E.; Heintz N. (2005) Large-scale genomic approaches to brain development and circuitry. Annu. Rev. Neurosci. 28, 89–108. [DOI] [PubMed] [Google Scholar]
- Luo L.; Callaway E. M.; Svoboda K. (2008) Genetic dissection of neural circuits. Neuron 57(5), 634–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Bernstein J. G.; Boyden E. S. (2011) Optogenetic tools for analyzing the neural circuits of behavior. Trends Cognit. Sci. 15(12), 592–600. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Fenno L.; Yizhar O.; Deisseroth K. (2011) The development and application of optogenetics. Annu. Rev. Neurosci. 34, 389–412. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Moglich A.; Moffat K. (2010) Engineered photoreceptors as novel optogenetic tools. Photochem. Photobiol. Sci. 9(10), 1286–1300. [DOI] [PubMed] [Google Scholar]
- a Boyden E. S.; Zhang F.; Bamberg E.; Nagel G.; Deisseroth K. (2005) Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8(9), 1263–1268. [DOI] [PubMed] [Google Scholar]; b Li X.; Gutierrez D. V.; Hanson M. G.; Han J.; Mark M. D.; Chiel H.; Hegemann P.; Landmesser L. T.; Herlitze S. (2005) Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc. Natl. Acad. Sci. U.S.A. 102(49), 17816–17821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner H. A.; Lein E. S.; Callaway E. M. (2002) A genetic method for selective and quickly reversible silencing of mammalian neurons. J. Neurosci. 22(13), 5287–5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgul N.; Weise C.; Kreienkamp H. J.; Richter D. (1999) Reverse physiology in Drosophila: Identification of a novel allatostatin-like neuropeptide and its cognate receptor structurally related to the mammalian somatostatin/galanin/opioid receptor family. EMBO J. 18(21), 5892–5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan E. M.; Yamaguchi Y.; Horwitz G. D.; Gosgnach S.; Lein E. S.; Goulding M.; Albright T. D.; Callaway E. M. (2006) Selective and quickly reversible inactivation of mammalian neurons in vivo using the Drosophila allatostatin receptor. Neuron 51(2), 157–170. [DOI] [PubMed] [Google Scholar]
- a Li P.; Slimko E. M.; Lester H. A. (2002) Selective elimination of glutamate activation and introduction of fluorescent proteins into a Caenorhabditis elegans chloride channel. FEBS Lett. 528(1–3), 77–82. [DOI] [PubMed] [Google Scholar]; b Slimko E. M.; Lester H. A. (2003) Codon optimization of Caenorhabditis elegans GluCl ion channel genes for mammalian cells dramatically improves expression levels. J. Neurosci. Methods 124(1), 75–81. [DOI] [PubMed] [Google Scholar]
- Lerchner W.; Xiao C.; Nashmi R.; Slimko E. M.; van Trigt L.; Lester H. A.; Anderson D. J. (2007) Reversible silencing of neuronal excitability in behaving mice by a genetically targeted, ivermectin-gated Cl- channel. Neuron 54(1), 35–49. [DOI] [PubMed] [Google Scholar]
- Armbruster B. N.; Li X.; Pausch M. H.; Herlitze S.; Roth B. L. (2007) Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc. Natl. Acad. Sci. U.S.A. 104(12), 5163–5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coward P.; Wada H. G.; Falk M. S.; Chan S. D.; Meng F.; Akil H.; Conklin B. R. (1998) Controlling signaling with a specifically designed Gi-coupled receptor. Proc. Natl. Acad. Sci. U.S.A. 95(1), 352–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S.; Rogan S. C.; Roth B. L. (2010) Directed molecular evolution of DREADDs: A generic approach to creating next-generation RASSLs. Nat. Protoc. 5(3), 561–573. [DOI] [PubMed] [Google Scholar]
- Alexander G. M.; Rogan S. C.; Abbas A. I.; Armbruster B. N.; Pei Y.; Allen J. A.; Nonneman R. J.; Hartmann J.; Moy S. S.; Nicolelis M. A.; McNamara J. O.; Roth B. L. (2009) Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. Neuron 63(1), 27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Eisele J. L.; Bertrand S.; Galzi J. L.; Devillers-Thiery A.; Changeux J. P.; Bertrand D. (1993) Chimaeric nicotinic-serotonergic receptor combines distinct ligand binding and channel specificities. Nature 366(6454), 479–483. [DOI] [PubMed] [Google Scholar]; b Grutter T.; de Carvalho L. P.; Dufresne V.; Taly A.; Edelstein S. J.; Changeux J. P. (2005) Molecular tuning of fast gating in pentameric ligand-gated ion channels. Proc. Natl. Acad. Sci. U.S.A. 102(50), 18207–18212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caterina M. J.; Schumacher M. A.; Tominaga M.; Rosen T. A.; Levine J. D.; Julius D. (1997) The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature 389(6653), 816–824. [DOI] [PubMed] [Google Scholar]
- Arenkiel B. R.; Klein M. E.; Davison I. G.; Katz L. C.; Ehlers M. D. (2008) Genetic control of neuronal activity in mice conditionally expressing TRPV1. Nat. Methods 5(4), 299–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guler A. D.; Rainwater A.; Parker J. G.; Jones G. L.; Argilli E.; Arenkiel B. R.; Ehlers M. D.; Bonci A.; Zweifel L. S.; Palmiter R. D. (2012) Transient activation of specific neurons in mice by selective expression of the capsaicin receptor. Nat. Commun. 3, 746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin D.; Madsen D.; Kahn-Kirby A.; Peckol E.; Moulder G.; Barstead R.; Maricq A.; Bargmann C. (2002) Combinatorial expression of TRPV channel proteins defines their sensory functions and subcellular localization in C. elegans neurons. Neuron 35(2), 307–318. [DOI] [PubMed] [Google Scholar]
- Wulff P.; Goetz T.; Leppa E.; Linden A. M.; Renzi M.; Swinny J. D.; Vekovischeva O. Y.; Sieghart W.; Somogyi P.; Korpi E. R.; Farrant M.; Wisden W. (2007) From synapse to behavior: Rapid modulation of defined neuronal types with engineered GABAA receptors. Nat. Neurosci. 10(7), 923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols C. D.; Roth B. L. (2009) Engineered G-protein coupled receptors are powerful tools to investigate biological processes and behaviors. Front. Mol. Neurosci. 2, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Redfern C. H.; Degtyarev M. Y.; Kwa A. T.; Salomonis N.; Cotte N.; Nanevicz T.; Fidelman N.; Desai K.; Vranizan K.; Lee E. K.; Coward P.; Shah N.; Warrington J. A.; Fishman G. I.; Bernstein D.; Baker A. J.; Conklin B. R. (2000) Conditional expression of a Gi-coupled receptor causes ventricular conduction delay and a lethal cardiomyopathy. Proc. Natl. Acad. Sci. U.S.A. 97(9), 4826–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Ma’ayan A.; Jenkins S. L.; Barash A.; Iyengar R. (2009) Neuro2A differentiation by G alpha i/o pathway. Sci. Signaling 2(54), cm1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Keramidas A.; Moorhouse A. J.; Schofield P. R.; Barry P. H. (2004) Ligand-gated ion channels: mechanisms underlying ion selectivity. Prog. Biophys. Mol. Biol. 86(2), 161–204. [DOI] [PubMed] [Google Scholar]; b Jensen M. L.; Schousboe A.; Ahring P. K. (2005) Charge selectivity of the Cys-loop family of ligand-gated ion channels. J. Neurochem. 92(2), 217–225. [DOI] [PubMed] [Google Scholar]; c Thompson A. J.; Lester H. A.; Lummis S. C. (2010) The structural basis of function in Cys-loop receptors. Q. Rev. Biophys. 43(4), 449–499. [DOI] [PubMed] [Google Scholar]
- Pei Y.; Rogan S. C.; Yan F.; Roth B. L. (2008) Engineered GPCRs as tools to modulate signal transduction. Physiology (Bethesda) 23, 313–321. [DOI] [PubMed] [Google Scholar]
- Haubensak W.; Kunwar P. S.; Cai H.; Ciocchi S.; Wall N. R.; Ponnusamy R.; Biag J.; Dong H. W.; Deisseroth K.; Callaway E. M.; Fanselow M. S.; Luthi A.; Anderson D. J. (2010) Genetic dissection of an amygdala microcircuit that gates conditioned fear. Nature 468(7321), 270–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Ma D.; Zerangue N.; Lin Y. F.; Collins A.; Yu M.; Jan Y. N.; Jan L. Y. (2001) Role of ER export signals in controlling surface potassium channel numbers. Science 291(5502), 316–319. [DOI] [PubMed] [Google Scholar]; b Gradinaru V.; Thompson K. R.; Deisseroth K. (2008) eNpHR: A Natronomonas halorhodopsin enhanced for optogenetic applications. Brain Cell Biol. 36(1–4), 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Srinivasan R.; Pantoja R.; Moss F. J.; Mackey E. D.; Son C. D.; Miwa J.; Lester H. A. (2011) Nicotine up-regulates alpha4beta2 nicotinic receptors and ER exit sites via stoichiometry-dependent chaperoning. J. Gen. Physiol. 137(1), 59–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Gradinaru V.; Thompson K. R.; Zhang F.; Mogri M.; Kay K.; Schneider M. B.; Deisseroth K. (2007) Targeting and readout strategies for fast optical neural control in vitro and in vivo. J. Neurosci. 27(52), 14231–14238. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Dong S.; Allen J. A.; Farrell M.; Roth B. L. (2010) A chemical-genetic approach for precise spatio-temporal control of cellular signaling. Mol. BioSyst. 6(8), 1376–1380. [DOI] [PubMed] [Google Scholar]; c Xu J.; Zhu Y.; Heinemann S. F. (2006) Identification of sequence motifs that target neuronal nicotinic receptors to dendrites and axons. J. Neurosci. 26(38), 9780–9793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Adelsberger H.; Lepier A.; Dudel J. (2000) Activation of rat recombinant alpha(1)beta(2)gamma(2S) GABA(A) receptor by the insecticide ivermectin. Eur. J. Pharmacol. 394(2–3), 163–170. [DOI] [PubMed] [Google Scholar]; b Shan Q.; Haddrill J. L.; Lynch J. W. (2001) Ivermectin, an unconventional agonist of the glycine receptor chloride channel. J. Biol. Chem. 276(16), 12556–12564. [DOI] [PubMed] [Google Scholar]; c Krause R. M.; Buisson B.; Bertrand S.; Corringer P. J.; Galzi J. L.; Changeux J. P.; Bertrand D. (1998) Ivermectin: A positive allosteric effector of the alpha7 neuronal nicotinic acetylcholine receptor. Mol. Pharmacol. 53(2), 283–294. [DOI] [PubMed] [Google Scholar]; d Khakh B. S.; Proctor W. R.; Dunwiddie T. V.; Labarca C.; Lester H. A. (1999) Allosteric control of gating and kinetics at P2X(4) receptor channels. J. Neurosci. 19(17), 7289–7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Smart D.; Gunthorpe M. J.; Jerman J. C.; Nasir S.; Gray J.; Muir A. I.; Chambers J. K.; Randall A. D.; Davis J. B. (2000) The endogenous lipid anandamide is a full agonist at the human vanilloid receptor (hVR1). Br. J. Pharmacol. 129(2), 227–230. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Huang S. M.; Bisogno T.; Trevisani M.; Al-Hayani A.; De Petrocellis L.; Fezza F.; Tognetto M.; Petros T. J.; Krey J. F.; Chu C. J.; Miller J. D.; Davies S. N.; Geppetti P.; Walker J. M.; Di Marzo V. (2002) An endogenous capsaicin-like substance with high potency at recombinant and native vanilloid VR1 receptors. Proc. Natl. Acad. Sci. U.S.A. 99(12), 8400–8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehr M.; Hostick U.; Kyweriga M.; Tan A.; Weible A. P.; Wu H.; Wu W.; Callaway E. M.; Kentros C. (2009) Transgenic silencing of neurons in the mammalian brain by expression of the allatostatin receptor (AlstR). J. Neurophysiol. 102(4), 2554–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalink K.; Moolenaar W. H. (2010) G protein-coupled receptors: The inside story. Bioessays 32(1), 13–16. [DOI] [PubMed] [Google Scholar]
- a Gunthorpe M. J.; Peters J. A.; Gill C. H.; Lambert J. J.; Lummis S. C. (2000) The 4′lysine in the putative channel lining domain affects desensitization but not the single-channel conductance of recombinant homomeric 5-HT3A receptors. J. Physiol. 522(Pt 2), 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hu X. Q.; Sun H.; Peoples R. W.; Hong R.; Zhang L. (2006) An interaction involving an arginine residue in the cytoplasmic domain of the 5-HT3A receptor contributes to receptor desensitization mechanism. J. Biol. Chem. 281(31), 21781–21788. [DOI] [PubMed] [Google Scholar]; c Giniatullin R.; Nistri A.; Yakel J. L. (2005) Desensitization of nicotinic ACh receptors: Shaping cholinergic signaling. Trends Neurosci. 28(7), 371–378. [DOI] [PubMed] [Google Scholar]; d Bouzat C.; Bartos M.; Corradi J.; Sine S. M. (2008) The interface between extracellular and transmembrane domains of homomeric Cys-loop receptors governs open-channel lifetime and rate of desensitization. J. Neurosci. 28(31), 7808–7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra D. P.; Nau C. (2003) Desensitization of capsaicin-activated currents in the vanilloid receptor TRPV1 is decreased by the cyclic AMP-dependent protein kinase pathway. J. Biol. Chem. 278(50), 50080–50090. [DOI] [PubMed] [Google Scholar]
- a Bunemann M.; Hosey M. M. (1999) G-protein coupled receptor kinases as modulators of G-protein signalling. J. Physiol. 517(Pt 1), 5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Bhave G.; Zhu W.; Wang H.; Brasier D. J.; Oxford G. S.; Gereau R. W. t. (2002) cAMP-dependent protein kinase regulates desensitization of the capsaicin receptor (VR1) by direct phosphorylation. Neuron 35(4), 721–731. [DOI] [PubMed] [Google Scholar]
- Scearce-Levie K.; Lieberman M. D.; Elliott H. H.; Conklin B. R. (2005) Engineered G protein coupled receptors reveal independent regulation of internalization, desensitization and acute signaling. BMC Biol. 3, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W.; Janczewski W. A.; Yang P.; Shao X. M.; Callaway E. M.; Feldman J. L. (2008) Silencing preBotzinger complex somatostatin-expressing neurons induces persistent apnea in awake rat. Nat. Neurosci. 11(5), 538–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Gosgnach S.; Lanuza G. M.; Butt S. J.; Saueressig H.; Zhang Y.; Velasquez T.; Riethmacher D.; Callaway E. M.; Kiehn O.; Goulding M. (2006) V1 spinal neurons regulate the speed of vertebrate locomotor outputs. Nature 440(7081), 215–219. [DOI] [PubMed] [Google Scholar]; b Zhang Y.; Narayan S.; Geiman E.; Lanuza G. M.; Velasquez T.; Shanks B.; Akay T.; Dyck J.; Pearson K.; Gosgnach S.; Fan C. M.; Goulding M. (2008) V3 spinal neurons establish a robust and balanced locomotor rhythm during walking. Neuron 60(1), 84–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson S. M.; Eskenazi D.; Ishikawa M.; Wanat M. J.; Phillips P. E.; Dong Y.; Roth B. L.; Neumaier J. F. (2011) Transient neuronal inhibition reveals opposing roles of indirect and direct pathways in sensitization. Nat. Neurosci. 14(1), 22–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray R. S.; Corcoran A. E.; Brust R. D.; Kim J. C.; Richerson G. B.; Nattie E.; Dymecki S. M. (2011) Impaired respiratory and body temperature control upon acute serotonergic neuron inhibition. Science 333(6042), 637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner A. R.; Rowland D. C.; Hwang S. Y.; Baumgaertel K.; Roth B. L.; Kentros C.; Mayford M. (2012) Generation of a synthetic memory trace. Science 335(6075), 1513–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K.; Suzuki M.; Mieda M.; Tsujino N.; Roth B.; Sakurai T. (2011) Pharmacogenetic modulation of orexin neurons alters sleep/wakefulness states in mice. PLoS One 6(5), e20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse A. C.; Hu J.; Pan A. C.; Arlow D. H.; Rosenbaum D. M.; Rosemond E.; Green H. F.; Liu T.; Chae P. S.; Dror R. O.; Shaw D. E.; Weis W. I.; Wess J.; Kobilka B. K. (2012) Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature 482(7386), 552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbs R. E.; Gouaux E. (2011) Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 474(7349), 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S. (2012) Membrane Proteins of Known 3D Structure http://blanco.biomol.uci.edu/mpstruc/listAll/list. [Google Scholar]
- a Romero P. A.; Arnold F. H. (2009) Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 10(12), 866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Guntas G.; Mansell T. J.; Kim J. R.; Ostermeier M. (2005) Directed evolution of protein switches and their application to the creation of ligand-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 102(32), 11224–11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor D. L. Jr. (2009) Searching for interesting channels: Pairing selection and molecular evolution methods to study ion channel structure and function. Mol. BioSyst. 5(8), 802–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone M. N.; Arnold F. H. (2007) Engineering by homologous recombination: Exploring sequence and function within a conserved fold. Curr. Opin. Struct. Biol. 17(4), 454–459. [DOI] [PubMed] [Google Scholar]
- Gaveriaux-Ruff C.; Kieffer B. L. (2007) Conditional gene targeting in the mouse nervous system: Insights into brain function and diseases. Pharmacol. Ther. 113(3), 619–634. [DOI] [PubMed] [Google Scholar]
- Gossen M.; Bujard H. (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. U.S.A. 89(12), 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M.; Freundlieb S.; Bender G.; Muller G.; Hillen W.; Bujard H. (1995) Transcriptional activation by tetracyclines in mammalian cells. Science 268(5218), 1766–1769. [DOI] [PubMed] [Google Scholar]
- Feil R.; Brocard J.; Mascrez B.; LeMeur M.; Metzger D.; Chambon P. (1996) Ligand-activated site-specific recombination in mice. Proc. Natl. Acad. Sci. U.S.A. 93(20), 10887–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymecki S. M.; Kim J. C. (2007) Molecular neuroanatomy’s “Three Gs”: A primer. Neuron 54(1), 17–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Xu J.; Pierson T.; O’Malley B. W.; Tsai S. Y. (1997) Positive and negative regulation of gene expression in eukaryotic cells with an inducible transcriptional regulator. Gene Ther. 4(5), 432–441. [DOI] [PubMed] [Google Scholar]
- Logie C.; Stewart A. F. (1995) Ligand-regulated site-specific recombination. Proc. Natl. Acad. Sci. U.S.A. 92(13), 5940–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellendonk C.; Tronche F.; Casanova E.; Anlag K.; Opherk C.; Schutz G. (1999) Inducible site-specific recombination in the brain. J. Mol. Biol. 285(1), 175–182. [DOI] [PubMed] [Google Scholar]
- Brocard J.; Feil R.; Chambon P.; Metzger D. (1998) A chimeric Cre recombinase inducible by synthetic,but not by natural ligands of the glucocorticoid receptor. Nucleic Acids Res. 26(17), 4086–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loew R.; Heinz N.; Hampf M.; Bujard H.; Gossen M. (2010) Improved Tet-responsive promoters with minimized background expression. BMC Biotechnol. 10, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Zhou X.; Vink M.; Klaver B.; Berkhout B.; Das A. T. (2006) Optimization of the Tet-On system for regulated gene expression through viral evolution. Gene Ther. 13(19), 1382–1390. [DOI] [PubMed] [Google Scholar]; b Urlinger S.; Baron U.; Thellmann M.; Hasan M. T.; Bujard H.; Hillen W. (2000) Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl. Acad. Sci. U.S.A. 97(14), 7963–7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Joyner A. L.; Zervas M. (2006) Genetic inducible fate mapping in mouse: Establishing genetic lineages and defining genetic neuroanatomy in the nervous system. Dev. Dyn. 235(9), 2376–2385. [DOI] [PubMed] [Google Scholar]; b Feil R. (2007) Conditional somatic mutagenesis in the mouse using site-specific recombinases. Handb. Exp. Pharmacol. 178, 3–28. [DOI] [PubMed] [Google Scholar]; c Hayashi S.; McMahon A. P. (2002) Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol. 244(2), 305–318. [DOI] [PubMed] [Google Scholar]
- Andersson H.; Alestig K. (1976) The penetration of doxycycline into CSF. Scand. J. Infect. Dis., Suppl. 9, 17–19. [PubMed] [Google Scholar]
- Agwuh K. N.; MacGowan A. (2006) Pharmacokinetics and pharmacodynamics of the tetracyclines including glycylcyclines. J. Antimicrob. Chemother. 58(2), 256–265. [DOI] [PubMed] [Google Scholar]
- Hasan M. T.; Schonig K.; Berger S.; Graewe W.; Bujard H. (2001) Long-term, noninvasive imaging of regulated gene expression in living mice. Genesis 29(3), 116–122. [DOI] [PubMed] [Google Scholar]
- a Wieland M.; Fussenegger M. (2010) Ligand-dependent regulatory RNA parts for Synthetic Biology in eukaryotes. Curr. Opin. Biotechnol. 21(6), 760–765. [DOI] [PubMed] [Google Scholar]; b Isaacs F. J.; Dwyer D. J.; Collins J. J. (2006) RNA synthetic biology. Nat. Biotechnol. 24(5), 545–554. [DOI] [PubMed] [Google Scholar]; c Chang A. L.; Wolf J. J.; Smolke C. D. (2012) Synthetic RNA switches as a tool for temporal and spatial control over gene expression. Curr. Opin. Biotechnol. 10.1016/j.copbio.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen L.; Svendsen J.; Lee J. S.; Gray J. T.; Magnier M.; Baba T.; D’Amato R. J.; Mulligan R. C. (2004) Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature 431(7007), 471–476. [DOI] [PubMed] [Google Scholar]
- Spencer D. M.; Wandless T. J.; Schreiber S. L.; Crabtree G. R. (1993) Controlling signal transduction with synthetic ligands. Science 262(5136), 1019–1024. [DOI] [PubMed] [Google Scholar]
- Clackson T.; Yang W.; Rozamus L. W.; Hatada M.; Amara J. F.; Rollins C. T.; Stevenson L. F.; Magari S. R.; Wood S. A.; Courage N. L.; Lu X.; Cerasoli F. Jr.; Gilman M.; Holt D. A. (1998) Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc. Natl. Acad. Sci. U.S.A. 95(18), 10437–10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Banaszynski L. A.; Wandless T. J. (2006) Conditional control of protein function. Chem. Biol. 13(1), 11–21. [DOI] [PubMed] [Google Scholar]; b Fegan A.; White B.; Carlson J. C.; Wagner C. R. (2010) Chemically controlled protein assembly: Techniques and applications. Chem Rev 110(6), 3315–3336. [DOI] [PubMed] [Google Scholar]
- Belshaw P. J.; Ho S. N.; Crabtree G. R.; Schreiber S. L. (1996) Controlling protein association and subcellular localization with a synthetic ligand that induces heterodimerization of proteins. Proc. Natl. Acad. Sci. U.S.A. 93(10), 4604–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayle J. H.; Grimley J. S.; Stankunas K.; Gestwicki J. E.; Wandless T. J.; Crabtree G. R. (2006) Rapamycin analogs with differential binding specificity permit orthogonal control of protein activity. Chem. Biol. 13(1), 99–107. [DOI] [PubMed] [Google Scholar]
- Kopytek S. J.; Standaert R. F.; Dyer J. C.; Hu J. C. (2000) Chemically induced dimerization of dihydrofolate reductase by a homobifunctional dimer of methotrexate. Chem. Biol. 7(5), 313–321. [DOI] [PubMed] [Google Scholar]
- Farrar M. A.; Olson S. H.; Perlmutter R. M. (2000) Coumermycin-induced dimerization of GyrB-containing fusion proteins. Methods Enzymol. 327, 421–429. [DOI] [PubMed] [Google Scholar]
- Clontech Inducible Systems. http://www.clontech.com/US/Products/Inducible_Systems/Inducible_Dimerization/iDimerize_Product_Overview?sitex=10020:22372:US.
- Karpova A. Y.; Tervo D. G.; Gray N. W.; Svoboda K. (2005) Rapid and reversible chemical inactivation of synaptic transmission in genetically targeted neurons. Neuron 48(5), 727–735. [DOI] [PubMed] [Google Scholar]
- Ostermeier M. (2009) Designing switchable enzymes. Curr. Opin. Struct. Biol. 19(4), 442–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski L. A.; Chen L. C.; Maynard-Smith L. A.; Ooi A. G.; Wandless T. J. (2006) A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 126(5), 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M.; Bjorklund T.; Lundberg C.; Kirik D.; Wandless T. J. (2010) A general chemical method to regulate protein stability in the mammalian central nervous system. Chem. Biol. 17(9), 981–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z.; Belforte J. E.; Lu Y.; Yabe Y.; Pickel J.; Smith C. B.; Je H. S.; Lu B.; Nakazawa K. (2010) eIF2alpha Phosphorylation-dependent translation in CA1 pyramidal cells impairs hippocampal memory consolidation without affecting general translation. J. Neurosci. 30(7), 2582–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slimko E. M.; McKinney S.; Anderson D. J.; Davidson N.; Lester H. A. (2002) Selective electrical silencing of mammalian neurons in vitro by the use of invertebrate ligand-gated chloride channels. J. Neurosci. 22(17), 7373–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furr B. J.; Jordan V. C. (1984) The pharmacology and clinical uses of tamoxifen. Pharmacol. Ther. 25(2), 127–205. [DOI] [PubMed] [Google Scholar]