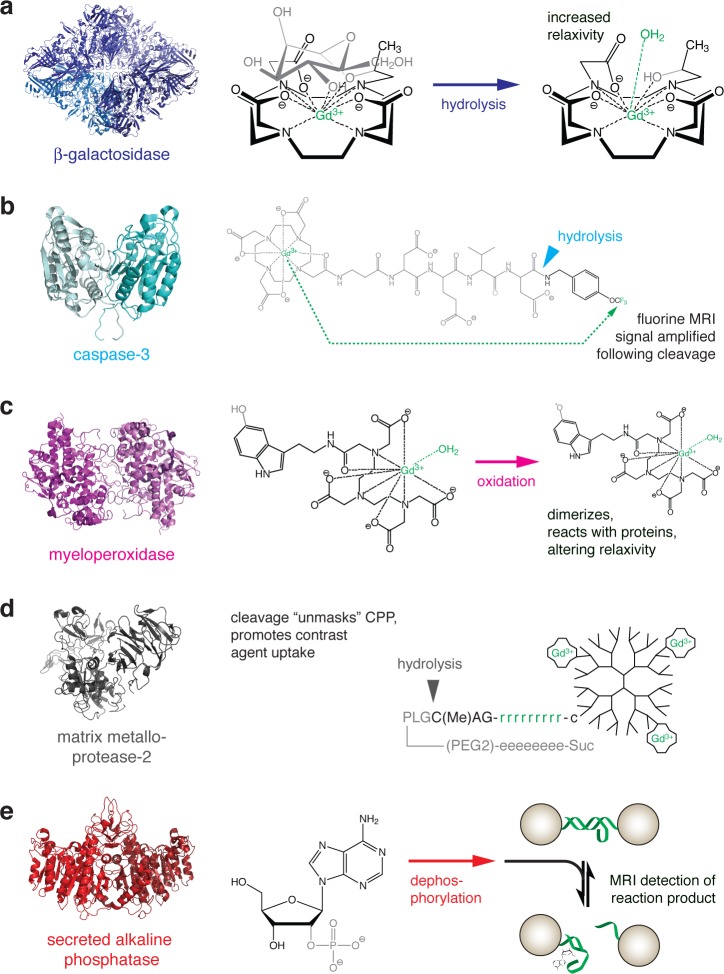
Figure 2.
Strategies for MRI-based detection of enzyme activity. For each example, the structure of the enzyme is shown at left, and the reaction catalyzed is shown at right. The chemical moieties most directly affected by the enzyme are shown in gray, and molecular components most directly responsible for MRI contrast are shown in green. (a) Gadolinium-containing substrate for β-galactosidase. Enzyme activity cleaves off a sugar moiety, increasing exposure of the gadolinium atom to interaction with a water molecule and consequently increasing T1 relaxivity.28 (b) Peptide-based probe for 19F MRI-based detection of caspase-3 activity. Prior to cleavage, the relaxation enhancement caused by the gadolinium chelate at left prevents detection of an 19F signal arising from the trifluoromethyl group at the right. Action of the enzyme cleaves the gadolinium-containing fragment and relieves the intramolecular relaxation effect, allowing an 19F signal to be detected.30 (c) Myeloperoxidase oxidizes the 5-hydroxy group of a serotonin-conjugated gadolinium chelate. The resulting free radical species tends to dimerize and react with proteins, resulting in compounds with longer τR and higher relaxivity.31 (d) A gadolinium-bearing dendrimer is conjugated to a peptide containing a poly-d-arginine cell penetrating domain, “masked” by an oppositely charged poly-d-glutamate domain. Action of matrix metalloprotease-2 or -9 cleaves the peptide, unmasking the polyarginine fragment and promoting accumulation of the contrast agent in nearby cells.37 (e) Detection of the reporter enzyme secreted alkaline phosphatase (SEAP) is performed using a nanoparticle-based T2 MRI sensor that detects adenosine, a product of SEAP-mediated dephosphorylation of 2′-adenosine monophosphate (left). Removal of adenosine by transport or further enzymatic processes reverses the contrast change mediated by the sensor.40