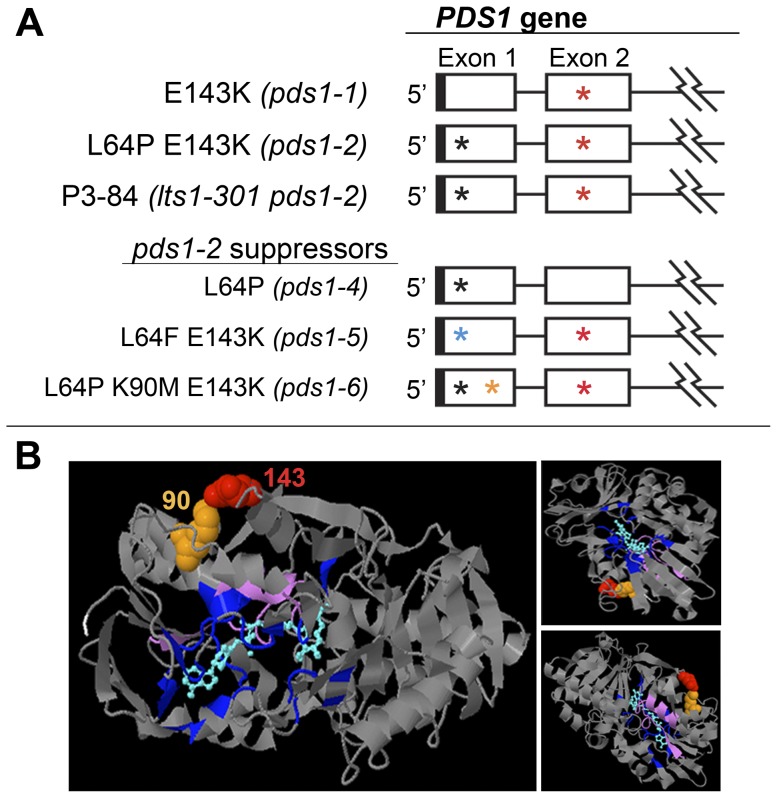
Figure 10. Analysis of intragenic suppressors of pds1-2 mutants.
A). Schematic depiction of pds1-1, P3-84, pds1-2, and pds1-2 suppressors (pds1-4, pds1-5, and pds1-6). Cartoon of the C. reinhardtii PDS gene showing only exons one and two. Jagged lines indicate a partial depiction of the PDS gene. UTRs are indicated by solid boxes, exons by open boxes and introns by lines. Asterisks mark positions of mutations in pds1-1, pds1-2, P3-84 and pds1-2 suppressor mutants. Black and blue asterisks represent mutations L64P and L64F, respectively. Orange asterisk in exon 1 signifies location of the K90M mutation, and red asterisks in exon 2 signify the E143K mutation. B). 3DLigandSite structural prediction of the C. reinhardtii PDS protein showing positions of amino acid residues mutated in pds1-1 and pds1-6 mutants, E143K and K90M, respectively. L64P and L64F were not mapped because the first 71 amino acids of the N-terminus had no structural prediction. Ligand and wild-type amino acids corresponding to mutated residues were colored as follows: position 90 (spacefilling, orange); position 143 (spacefilling, red); ligand [NAD(P)/FAD] (cyan); predicted ligand binding sites (indigo); start of predicted N-terminus (amino acid residue 72, white); and carotenoid binding site proposed by Armstrong et al. (amino acid residues 492–517, lavender). Three different perspectives of the predicted structure are shown.