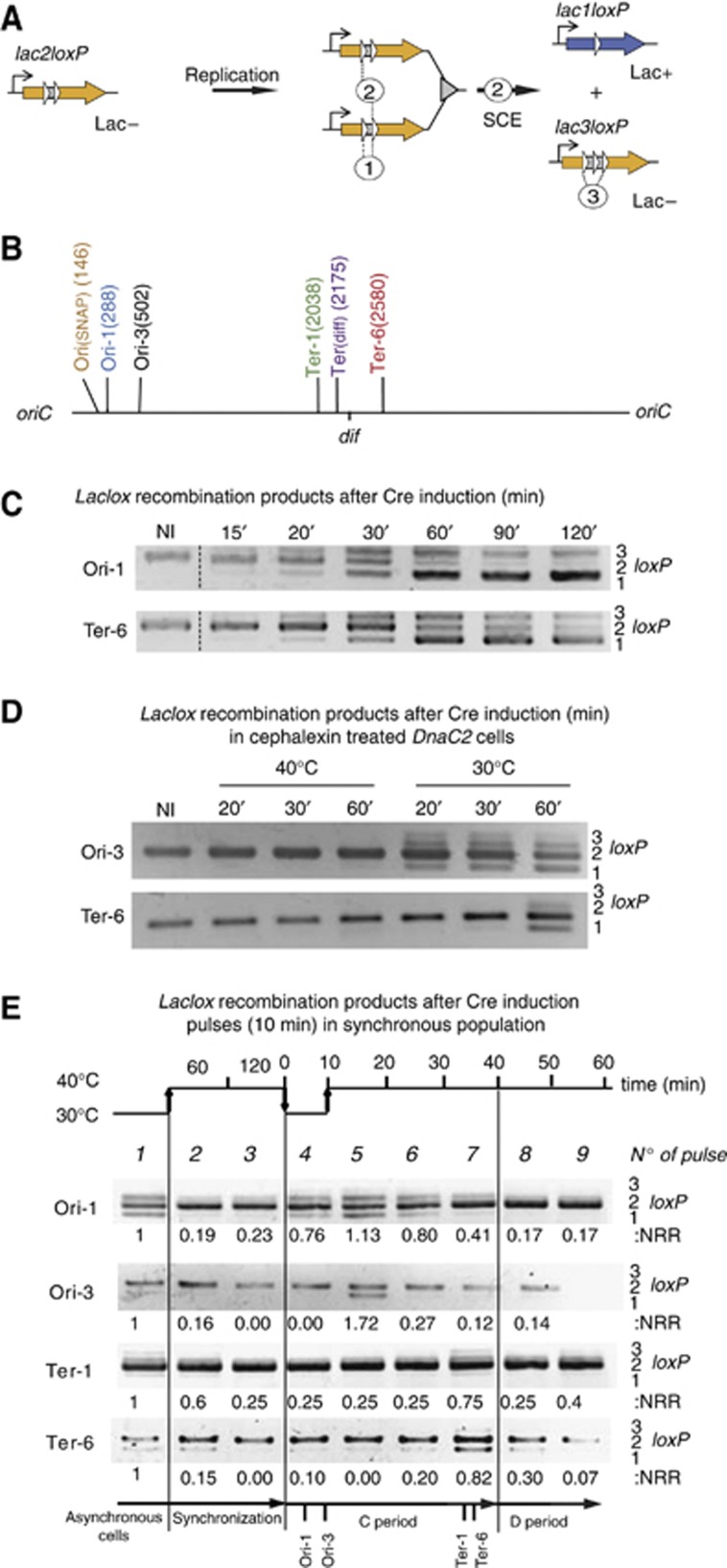
Figure 1.
The LacloxP assay revealed sister chromatid interactions. (A) The LacloxP construct consisted of the lacZ gene (orange arrow) under a constitutive promoter that was interrupted by two directly repeated loxP sites (white arrows). This construct conferred the Lac− phenotype. The intramolecular recombination event (noted 1) was prevented by the proximity between the two loxP sites. After replication, intermolecular recombination occurred between sister chromatids (noted 2) when Cre was produced. This recombination forms the lac1loxP product (blue arrow), conferring the Lac+ phenotype. The intramolecular recombination between the first and third loxP sites of lac3loxP product is noted event 3. (B) Map of the E. coli chromosome with the position of the six loci tested in this study labeled. The positions are indicated in kb from the oriC. (C) Recombination events were revealed using PCR amplification that detected the substrate, lac2loxP (406 bp), and products, lac1loxP (351 bp) and lac3loxP (460 bp). The gel shows PCR amplification obtained on total DNA sample extracted from MG1655 strains containing the LacloxP system at the Ori-1 or Ter-6 loci before (NI) and after Cre induction for the indicated time. (D) LacloxP recombination happens on replicating chromosomes but not fully segregated chromosomes. Recombination events were revealed using PCR amplification following Cre induction in the MG1655dnaC2 strain. Replication initiation was blocked by a 2-h shift to a non-permissive temperature (40°C) simultaneously cytokinesis was prohibited by the addition of cephalexin (10 μg/ml). In these conditions the cells filamented and accumulated segregated nucleoids (data not shown). Cre induction was performed at 40°C for 20, 30 or 60 min, or following replication initiation after a downshift from 40 to 30°C for 20, 30 and 60 min. (E) LacloxP recombination follows the replication forks. We used the dnaC2 thermosensitive allele to synchronize the population. Cells were grown at 30°C until OD 0.2 and were shifted to 40°C for two hours to allow the completion of ongoing replications. Replication was initiated by a 10 min shift at 30°C, and cells were returned to 40°C to avoid over-initiation. Nine pulses of 10 min Cre induction were performed and then the cells were immediately harvested and frozen in liquid nitrogen. Genomic DNA was extracted and used for PCR amplification, revealing the formation of LacloxP recombination products. The quantification of recombination is presented (NRR), it corresponds to the measure of the amount of lac1loxP plus lac3loxP compared to the amount of lac2loxP. The values were normalized to that observed for the asynchronous culture.