Figure 1.
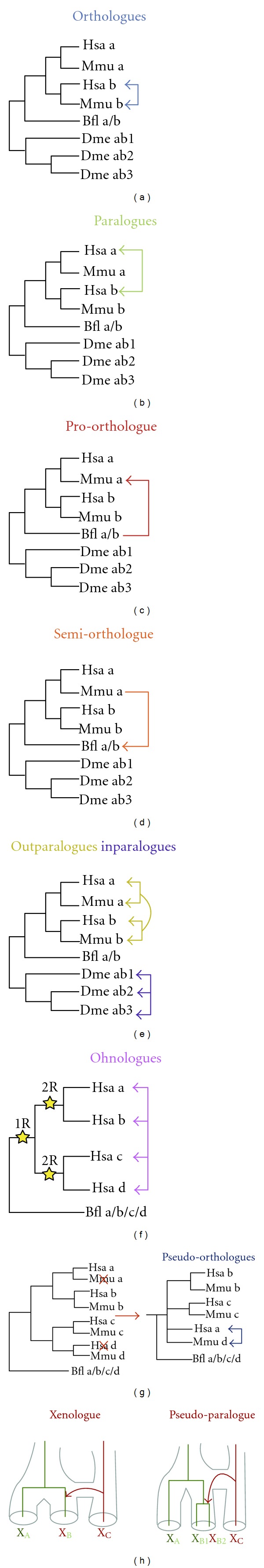
Overview of the current terminology. The different panels represent term(s) for duplicated genes. (a) Orthologues. The square blue arrows represent an orthologous relationship between the two genes. (b) Paralogues. The square green arrows represent paralogous relationships between the genes. (c) Proto-orthologue. The square red arrow represents the pro-orthologue relationship of gene a/b from Branchiostoma floridae to gene a from Mus musculus. (d) Semi-orthologue. The square orange arrow represents the semi-orthologous relationship of gene a of Mus musculus to gene a/b from Branchiostoma floridae. (e) Inparalogues and Outparalogues. The square yellow arrows represent the outparalogous relationship in which human and mouse a genes are outparalogous to human and mouse b genes. As a set, genes a and b from mouse and human represents coorthologues. The square purple arrows represent the inparalogous relationship between the genes which duplicated within this lineage. (f) Ohnologues. The square pink arrows delimit all the paralogues coming from WGD and the stars represent the duplication events. (g) Pseudo-orthologues. The square navy arrows represent the pseudo-orthologues. The red Xs represent lineage-specific gene losses. (h) Xenologues and Pseudo-paralogues. Species are represented by subindices A, B, and C, and the Xs represent the orthologous genes with their colouring designating the species of origin. All of the figures are adapted from [4–6]. Bfl: Branchiostoma floridae, Dme: Drosophila melanogaster, Hsa: Homo sapiens, and Mmu: Mus musculus.
