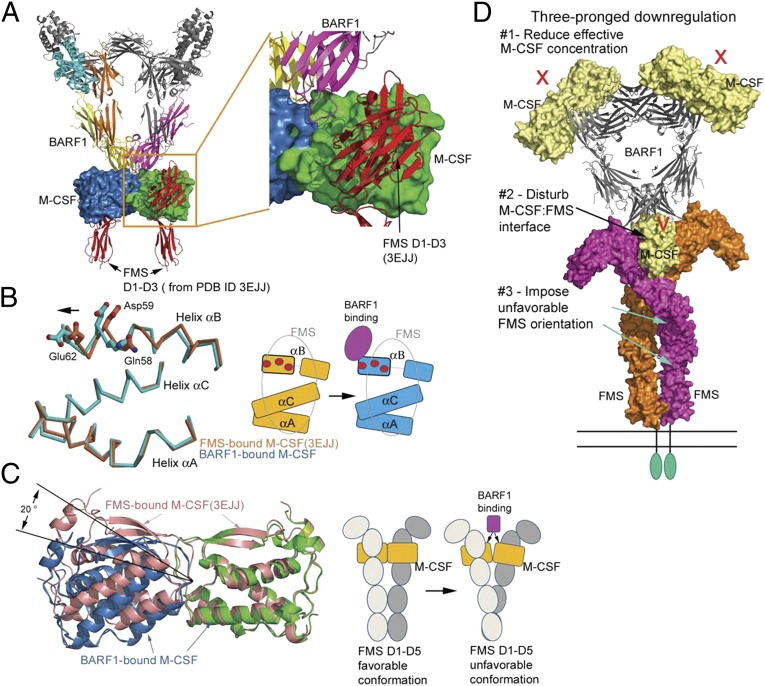
Fig. 4.
Mechanism of BARF1 down-regulation of M-CSF:FMS signaling. (A) Docking FMS to the BARF1:M-CSF complex, based on the previous M-CSF:FMS complex structure (PDB ID code 3EJJ), revealing that there is no overlap between BARF1 and FMS. (B) The conformational change of the M-CSF helix αB upon BARF1 binding. (Left) The helices of BARF1-bound M-CSF (cyan) are superimposed on the helices of FMS-bound M-CSF (salmon), revealing half of the helix αB shifting toward incoming BARF1. The M-CSF residues important for FMS binding are shown as balls and sticks. (Right) Of the three FMS-binding helices of M-CSF, only half of helix αB is shifted by BARF1, creating a partial mismatch with FMS (gray oval). The same set of FMS-binding residues is shown as red circles in the cartoon. (C) The bending of M-CSF dimer by BARF1 and the effect of the bending on FMS conformation. (Left) The BARF1-bound M-CSF dimer (blue and green) is overlaid on the FMS-bound M-CSF dimer (salmon) by superimposing one of their M-CSF protomers. This superimposition reveals that BARF1 forces M-CSF to bend by 20° relative to the center of the dimer, compared with the FMS-bound state. (Right) The cartoons demonstrate the possible effect of such bending on the FMS conformation. (D) A model summarizing the three-pronged approach that BARF-1 may use to down-regulate M-CSF signaling mediated by FMS.