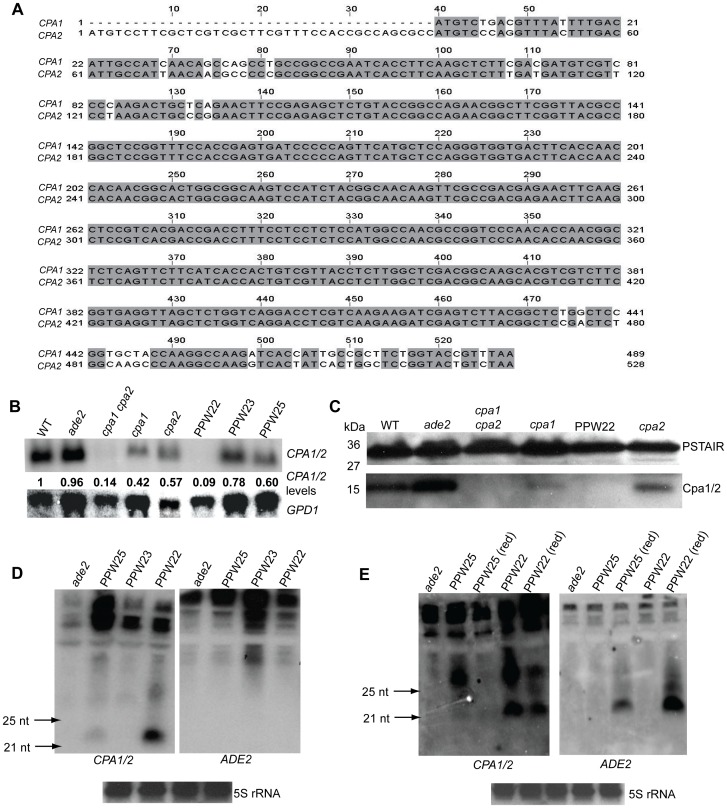
Figure 4. Silencing of the CPA1 and CPA2 genes in strains bearing a cpa1::ADE2 transgene array.
A) The CPA1 and CPA2 genes are highly conserved and share ∼93% nucleotide identity in the coding regions (introns are not depicted), including a region of 339 bp with 100% identity. Identical nucleotides are darkly shaded. B) Expression of CPA1 and CPA2 was examined by northern blot analysis. The full-length CPA1 gene was used as a probe and hybridizes with both the CPA1 and the CPA2 mRNAs. The GPD1 gene served as the loading control. The quantities of the CPA1/2 gene mRNA were determined by phosphorimager analysis and Image Quantifier 5.2 software. The quantity of CPA1/2 in each sample was compared with that in the wild-type strain (set as 1) after normalization with GPD1. C) Western blot detection of C. neoformans protein extracts indicates that the abundance of both the Cpa1 and Cpa2 proteins is reduced in the transgenic strain PPW22. Total proteins from the wild-type, ade2, cpa1, cpa2, cpa1 cpa2, and PPW22 strains were separated on a 4–20% SDS-PAGE gel, blotted, and incubated with anti-Cpa1 antiserum. The same membrane was stripped and probed with anti-PSTAIR antibodies to detect the Cdc2 protein as a loading control. D) CPA1 siRNAs are present in the transformed strains exhibiting CsA resistance. Total RNAs were isolated and siRNAs were enriched and resolved in a 15% denaturing polyacrylamide TBE-urea gel and probed with a sense CPA1 probe. Northern analysis revealed that CPA1 siRNAs are highly abundant in transgenic strain PPW22, but much less abundant in PPW25. No siRNA was detected when the blot was probed with a 32P-labeled ADE2 sense probe. E) Abundant ADE2 siRNAs were only observed when RNA was extracted from red (ade−) colonies of the variegating strains (red/white). 5S rRNA was probed to serve as a loading control.