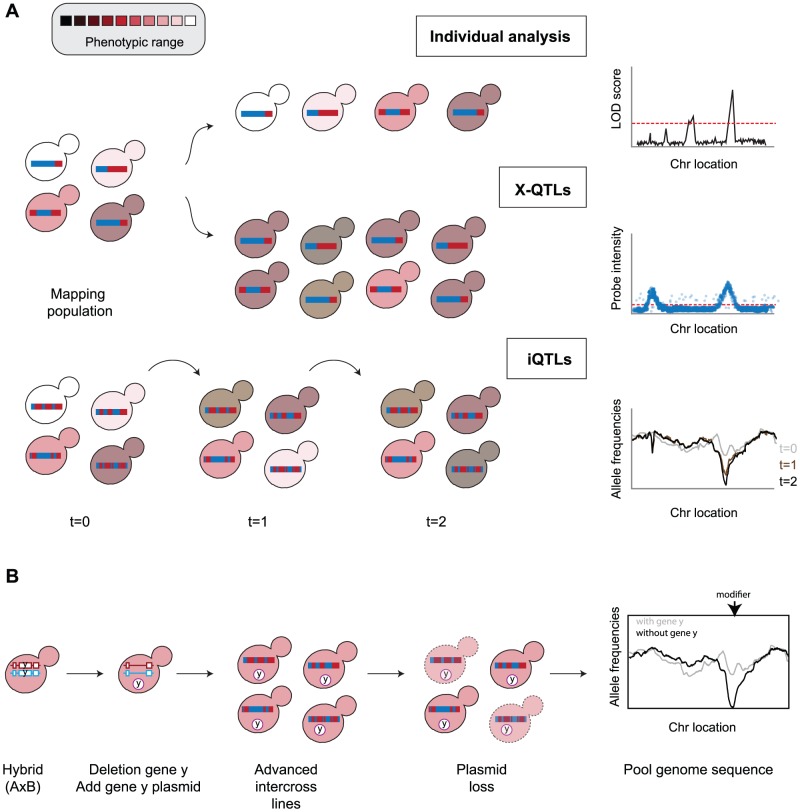
Figure 2. Mapping QTLs and modifiers.
(A) QTL mapping has evolved from the classical approach of individual segregant analysis to the X-QTLs and iQTLs approaches with higher mapping sensitivity and resolution. Analysis of time series data in iQTLs allows dynamic monitoring of allele frequency values [56]. (B) A possible approach to map genetic modifiers using iQTLs. A conditional essential gene, y, is deleted from its original chromosomal location and maintained on a plasmid. This hybrid is intercrossed multiple times to allow reshuffling of parental genomes. Upon loss of gene y, viability relies on the presence of genetic modifier/s, and allelic combinations that result in lethality (dashed cells) will decrease in allele frequency. These modifiers can be detected by comparing allele frequencies of the pool before and after the plasmid loss. When many modifiers are involved, the lethal combinations will be present in low frequency, making them difficult to detect. Further rounds of intercrosses, after loss of gene y, will allow reshuffling of alleles and the generation of more cells with unviable combinations.