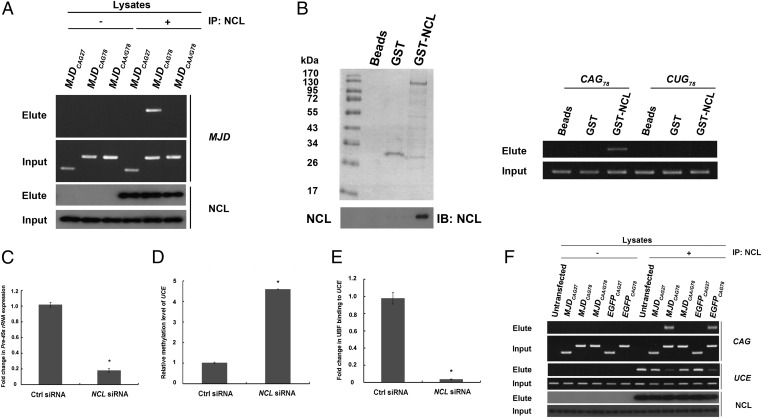
Fig. 3.
Physical interaction between expanded CAG RNAs and nucleolin compromised the binding of nucleolin to upstream control element. (A) Pull-down of MJDCAG RNA by means of NCL immunoprecipitation. “+” indicates that antibody was present in the immunoprecipitation reactions and “−” indicates that no antibody was included in the reactions. This experiment was repeated three times, and a representative gel is shown. (B) Direct physical interaction between NCL and expanded CAG RNA. Purified GST-NCL protein and in vitro transcribed RNAs (CAG78 and CUG78) were used in the binding reactions. Nonfusion GST protein was used as a negative control. Western blotting was performed to confirm the expression of the GST-NCL protein. This experiment was repeated four times, and a representative blot is shown. (C) Real-time PCR analysis of pre-45s rRNA expression levels in NCL siRNA-treated cells. Error bars represent ±SD. This experiment was repeated three times. (D) Real-time PCR analysis of UCE DNA methylation status in NCL siRNA-treated cells. Error bars represent ±SD. This experiment was repeated three times. (E) Coimmunoprecipitation of upstream binding factor (UBF) and upstream control element (UCE) in NCL siRNA-treated cells. Real-time PCR analysis was performed to determine UBF–UCE interaction. Error bars represent ±SD. This experiment was repeated three times. (F) Chromatin immunoprecipitation of NCL with expanded CAG RNAs and UCE in cells. “+” indicates that antibody was present in the immunoprecipitation reactions and “−” indicates that no antibody was included in the reactions. This experiment was repeated three times, and a representative gel is shown.