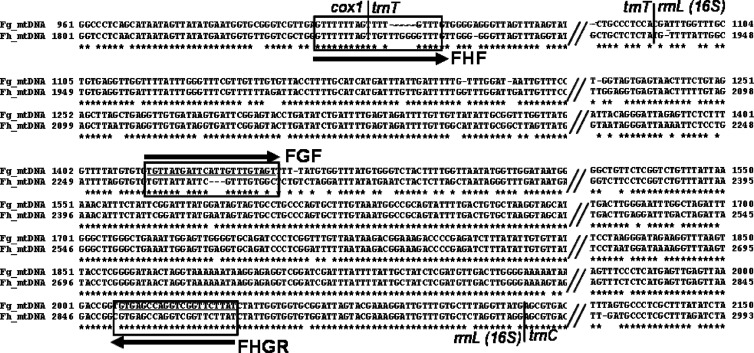
Fig 1.
Comparative alignment of the mitochondrial genome spanning cox1-trnT-rrnL sequences of F. gigantica and F. hepatica for their variability between two species in regions chosen (boxed) for the species primers of multiplex PCR (here referred to as duplex PCR). The upper line in each block is the nucleotide sequence of F. gigantica (FspT4, Vietnam) and the lower is that of F. hepatica (FhAUS, Australia). Alignment gaps are indicated by hyphens and omitted regions by double slashes. Borders between genes are indicated. The horizontal arrows and names indicate direction (sense and antisense) of the primers (underlined), including FHF (forward F. hepatica primer), FGF (forward F. gigantica primer), and FHGR (common reverse primer).